Create Ding et al. datasets - Mixture 3T3/HEK
Last updated: 2021-12-17
Checks: 7 0
Knit directory: sct2_revision/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210706) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 8afc486. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/raw_data/
Ignored: data/rds_filtered/
Ignored: data/rds_raw/
Ignored: data/sampled_counts/
Ignored: output/snakemake_output/
Untracked files:
Untracked: code/02_run_seurat_noclip.R
Untracked: code/07AA_deseq2_muscat_simulate.R
Untracked: code/07A_muscat_simulate.R
Untracked: code/07A_simulate_muscat.R
Untracked: code/07BB_deseq2_muscat_process.R
Untracked: code/07B_muscat_process.R
Untracked: code/07B_process_muscat.R
Untracked: code/08_run_presto.R
Untracked: code/17A_HEK_SS3_dropseq.Rmd
Untracked: code/17A_HEK_SS3_dropseq_files/
Untracked: code/17C_HEK_Quartzeseq2_dropseq.Rmd
Untracked: code/17C_HEK_Quartzeseq2_dropseq_files/
Untracked: code/17_HEK_SS3_ChromiumV3.Rmd
Untracked: code/17_HEK_SS3_ChromiumV3.nb.html
Untracked: code/17_HEK_SS3_ChromiumV3_files/
Untracked: code/AA_process_muscat.R
Untracked: code/BB_process_muscat.R
Untracked: code/DD_simulate_muscat.R
Untracked: code/EE_simulate_muscat.R
Untracked: code/XX_process_muscat.R
Untracked: code/XX_simulate_muscat.R
Untracked: code/YY_simulate_muscat.R
Untracked: code/ZZ_simulate_muscat.R
Untracked: code/kang_muscat.R
Untracked: code/prep_sce.R
Untracked: code/prep_sce_ss3_dropseq.R
Untracked: data/azimuth_predictions/
Untracked: junk/
Untracked: mamba_update_changes.txt
Untracked: output/11C_VST/
Untracked: output/AAmuscat_simulated/
Untracked: output/BBmuscat_simulated/
Untracked: output/CCmuscat_simulated/
Untracked: output/CD4_NK_downsampling_DE.rds
Untracked: output/DDmuscat_simulated/
Untracked: output/EEmuscat_simulated/
Untracked: output/KANGmuscat_simulated/
Untracked: output/NK_downsampling/
Untracked: output/XXmuscat_simulated/
Untracked: output/YYmuscat_simulated/
Untracked: output/ZZmuscat_simulated/
Untracked: output/figures/
Untracked: output/kang_prepsce.rds
Untracked: output/muscat_simulated/
Untracked: output/muscat_simulation/
Untracked: output/seu_sct2_sim.rds
Untracked: output/simulation_HEK_QuartzSeq2_Dropseq_downsampling/
Untracked: output/simulation_HEK_SS3_ChromiumV3_downsampling/
Untracked: output/simulation_HEK_SS3_Dropseq_downsampling/
Untracked: output/simulation_HEK_downsampling/
Untracked: output/simulation_NK_downsampling/
Untracked: output/ss3_dropseq_prepsim.rds
Untracked: output/tables/
Untracked: output/vargenes/
Untracked: snakemake/.snakemake/
Untracked: snakemake/Snakefile_noclip.smk
Untracked: snakemake/Snakefile_presto.smk
Untracked: snakemake/cluster.yaml
Untracked: snakemake/install_glm.R
Untracked: snakemake/jobscript.sh
Untracked: snakemake/jobscript_ncells.sh
Untracked: snakemake/local_run_downsampling.sh
Untracked: snakemake/local_run_glm.sh
Untracked: snakemake/local_run_ncells.sh
Untracked: snakemake/local_run_noclip.sh
Untracked: snakemake/local_run_presto.sh
Untracked: snakemake/local_run_time.sh
Untracked: snakemake/run_glm.sh
Untracked: snakemake/run_ncells.sh
Untracked: snakemake/sct2_revision_env.yml
Untracked: temp_figures/
Unstaged changes:
Deleted: analysis/04_PBMC68k.Rmd
Modified: code/02_run_seurat.R
Modified: code/03_run_vst2_downsample.R
Modified: code/04_run_vst_ncells.R
Modified: code/06_run_sct.R
Modified: data/datasets.csv
Modified: snakemake/Snakefile_downsampling.smk
Modified: snakemake/Snakefile_glm_seurat.smk
Modified: snakemake/Snakefile_metacell.smk
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/03A_Ding-Mixture-HEK-3T3.Rmd) and HTML (docs/03A_Ding-Mixture-HEK-3T3.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | d736ec8 | Saket Choudhary | 2021-07-07 | Build site. |
| Rmd | 400797a | Saket Choudhary | 2021-07-06 | workflowr::wflow_git_commit(all = TRUE) |
| html | 400797a | Saket Choudhary | 2021-07-06 | workflowr::wflow_git_commit(all = TRUE) |
suppressPackageStartupMessages({
library(Seurat)
library(ggplot2)
library(patchwork)
library(stringr)
library(httr)
library(XML)
})
set.seed(42)
theme_set(theme_classic())
FetchGEOFiles <- function(geo, download.dir = getwd(), download.files = FALSE, ...) {
geo <- trimws(toupper(geo))
geo_type <- substr(geo, 1, 3)
url.prefix <- "https://ftp.ncbi.nlm.nih.gov/geo/"
if (geo_type == "GSE") {
url.prefix <- paste0(url.prefix, "series/")
} else if (geo_type == "GSM") {
url.prefix <- paste0(url.prefix, "samples/")
} else if (geotype == "GPL") {
url.prefix <- paste0(url.prefix, "platform/")
}
geo_prefix <- paste0(substr(x = geo, start = 1, stop = nchar(geo) - 3), "nnn")
url <- paste0(url.prefix, geo_prefix, "/", geo, "/", "suppl", "/")
response <- GET(url = url)
html_parsed <- htmlParse(file = response)
links <- xpathSApply(doc = html_parsed, path = "//a/@href")
suppl_files <- as.character(grep(pattern = "^G", x = links, value = TRUE))
if (length(suppl_files) == 0) {
return(NULL)
}
file.url <- paste0(url, suppl_files)
file_list <- data.frame(filename = suppl_files, url = file.url)
if (download.files) {
names(file.url) <- suppl_files
download_file <- function(url, filename, ...) {
message(paste0("Downloading ", filename, " to ", download.dir))
download.file(url = url, destfile = file.path(download.dir, filename), mode = "wb", ...)
message("Done!")
}
lapply(seq_along(file.url), function(y, n, i) {
download_file(y[[i]], n[[i]], ...)
},
y = file.url, n = names(file.url)
)
}
return(file_list)
}download_dir <- here::here("data/raw_data/Ding")
dir.create(download_dir, showWarnings = F, recursive = T)
dir.create(here::here("data/rds_raw"), showWarnings = F, recursive = T)
geo_files <- FetchGEOFiles("GSE132044", download_dir, download.files = T)
geo_files filename
1 GSE132044_HEK293_PBMC_TPM_bulk.tsv.gz
2 GSE132044_NIH3T3_cortex_TPM_bulk.tsv.gz
3 GSE132044_cortex_mm10_cell.tsv.gz
4 GSE132044_cortex_mm10_count_matrix.mtx.gz
5 GSE132044_cortex_mm10_gene.tsv.gz
6 GSE132044_mixture_hg19_mm10_cell.tsv.gz
7 GSE132044_mixture_hg19_mm10_count_matrix.mtx.gz
8 GSE132044_mixture_hg19_mm10_gene.tsv.gz
9 GSE132044_pbmc_hg38_cell.tsv.gz
10 GSE132044_pbmc_hg38_count_matrix.mtx.gz
11 GSE132044_pbmc_hg38_gene.tsv.gz
url
1 https://ftp.ncbi.nlm.nih.gov/geo/series/GSE132nnn/GSE132044/suppl/GSE132044_HEK293_PBMC_TPM_bulk.tsv.gz
2 https://ftp.ncbi.nlm.nih.gov/geo/series/GSE132nnn/GSE132044/suppl/GSE132044_NIH3T3_cortex_TPM_bulk.tsv.gz
3 https://ftp.ncbi.nlm.nih.gov/geo/series/GSE132nnn/GSE132044/suppl/GSE132044_cortex_mm10_cell.tsv.gz
4 https://ftp.ncbi.nlm.nih.gov/geo/series/GSE132nnn/GSE132044/suppl/GSE132044_cortex_mm10_count_matrix.mtx.gz
5 https://ftp.ncbi.nlm.nih.gov/geo/series/GSE132nnn/GSE132044/suppl/GSE132044_cortex_mm10_gene.tsv.gz
6 https://ftp.ncbi.nlm.nih.gov/geo/series/GSE132nnn/GSE132044/suppl/GSE132044_mixture_hg19_mm10_cell.tsv.gz
7 https://ftp.ncbi.nlm.nih.gov/geo/series/GSE132nnn/GSE132044/suppl/GSE132044_mixture_hg19_mm10_count_matrix.mtx.gz
8 https://ftp.ncbi.nlm.nih.gov/geo/series/GSE132nnn/GSE132044/suppl/GSE132044_mixture_hg19_mm10_gene.tsv.gz
9 https://ftp.ncbi.nlm.nih.gov/geo/series/GSE132nnn/GSE132044/suppl/GSE132044_pbmc_hg38_cell.tsv.gz
10 https://ftp.ncbi.nlm.nih.gov/geo/series/GSE132nnn/GSE132044/suppl/GSE132044_pbmc_hg38_count_matrix.mtx.gz
11 https://ftp.ncbi.nlm.nih.gov/geo/series/GSE132nnn/GSE132044/suppl/GSE132044_pbmc_hg38_gene.tsv.gzMixture
mixture <- ReadMtx(mtx = "~/github/scRNA_NB_comparison/data/raw_data/Ding/GSE132044_mixture_hg19_mm10_count_matrix.mtx.gz",
cells = "~/github/scRNA_NB_comparison/data/raw_data/Ding/GSE132044_mixture_hg19_mm10_cell.tsv.gz",
features = "~/github/scRNA_NB_comparison/data/raw_data/Ding/GSE132044_mixture_hg19_mm10_gene.tsv.gz",
feature.column = 1
)mixture[1:5, 1:5]5 x 5 sparse Matrix of class "dgCMatrix"
Mixture1.Smart-seq2.p2_A4
hg19_ENSG00000000003_hg19_TSPAN6 .
hg19_ENSG00000000005_hg19_TNMD .
hg19_ENSG00000000419_hg19_DPM1 .
hg19_ENSG00000000457_hg19_SCYL3 .
hg19_ENSG00000000460_hg19_C1orf112 .
Mixture1.Smart-seq2.p2_A7
hg19_ENSG00000000003_hg19_TSPAN6 .
hg19_ENSG00000000005_hg19_TNMD .
hg19_ENSG00000000419_hg19_DPM1 .
hg19_ENSG00000000457_hg19_SCYL3 .
hg19_ENSG00000000460_hg19_C1orf112 .
Mixture1.Smart-seq2.p2_B1
hg19_ENSG00000000003_hg19_TSPAN6 270
hg19_ENSG00000000005_hg19_TNMD .
hg19_ENSG00000000419_hg19_DPM1 .
hg19_ENSG00000000457_hg19_SCYL3 .
hg19_ENSG00000000460_hg19_C1orf112 213
Mixture1.Smart-seq2.p1_A7
hg19_ENSG00000000003_hg19_TSPAN6 .
hg19_ENSG00000000005_hg19_TNMD .
hg19_ENSG00000000419_hg19_DPM1 .
hg19_ENSG00000000457_hg19_SCYL3 .
hg19_ENSG00000000460_hg19_C1orf112 .
Mixture1.Smart-seq2.p2_B12
hg19_ENSG00000000003_hg19_TSPAN6 .
hg19_ENSG00000000005_hg19_TNMD .
hg19_ENSG00000000419_hg19_DPM1 .
hg19_ENSG00000000457_hg19_SCYL3 .
hg19_ENSG00000000460_hg19_C1orf112 .replicate <- str_split_fixed(colnames(mixture), pattern = "\\.", n = 3)[, 1]
technology <- str_split_fixed(colnames(mixture), pattern = "\\.", n = 3)[, 2]
metadata <- data.frame(technology = technology, replicate = replicate)
rownames(metadata) <- colnames(mixture)
genes <- rownames(mixture)
species <- str_split_fixed(genes, pattern = "_", n = 4)[, 1]
hg19_genes <- species == "hg19"
mm10_genes <- species == "mm10"
mixture_hg19 <- mixture[hg19_genes, ]
mixture_mm10 <- mixture[mm10_genes, ]qplot(colSums(mixture_hg19), colSums(mixture_mm10))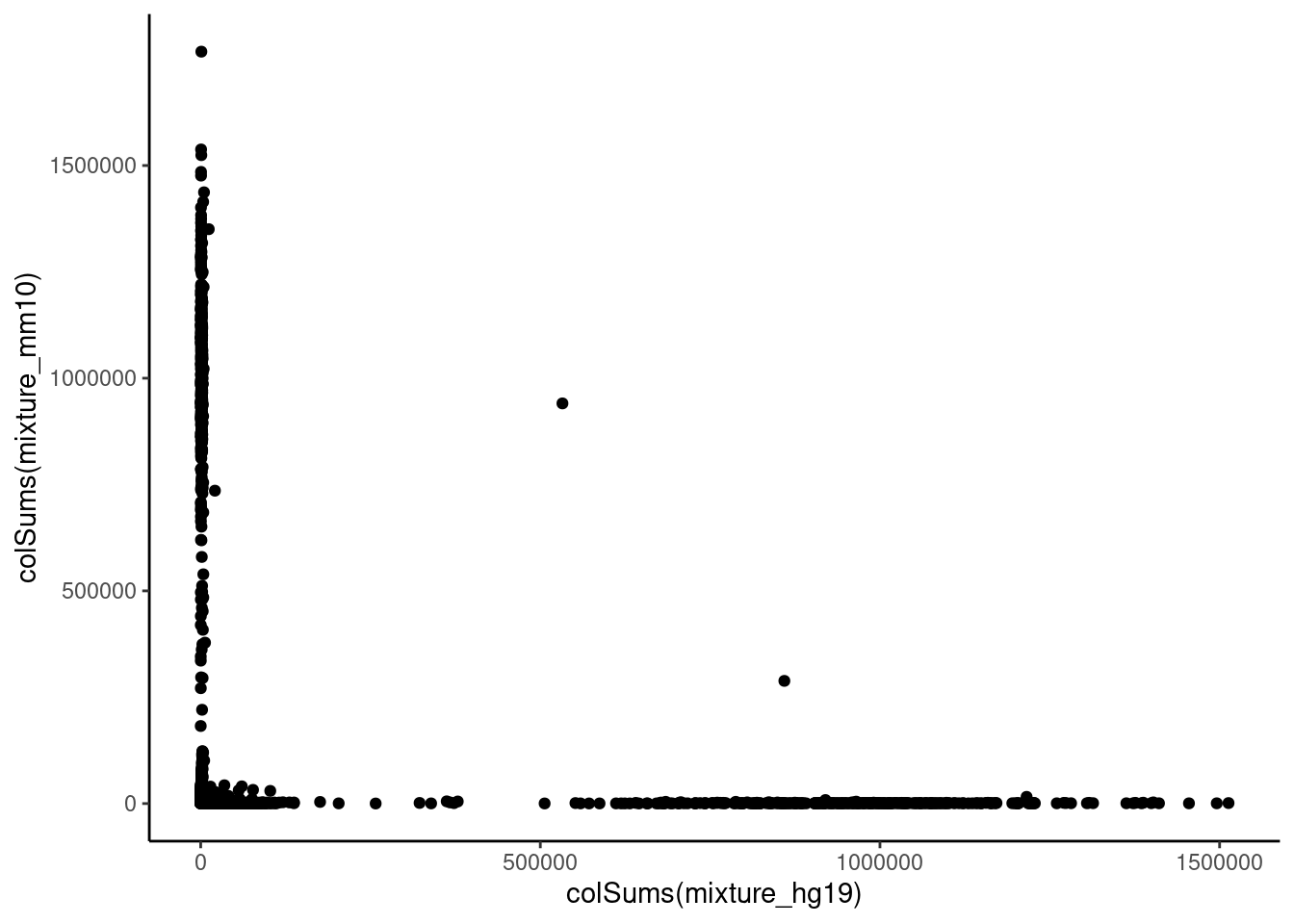
| Version | Author | Date |
|---|---|---|
| 400797a | Saket Choudhary | 2021-07-06 |
total_cell_umi <- data.frame(hg19_umi = colSums(mixture_hg19), mm10_umi = colSums(mixture_mm10))
total_cell_umi$hg19_over_mm10 <- total_cell_umi$hg19_umi / total_cell_umi$mm10_umi
human_cells <- rownames(total_cell_umi[total_cell_umi$hg19_over_mm10 >= 0.75, ])
mouse_cells <- rownames(total_cell_umi[total_cell_umi$hg19_over_mm10 < 0.25, ])
dim(total_cell_umi)[1] 27714 3length(human_cells)[1] 16163length(mouse_cells)[1] 11338mixture_hg19_humanonly <- mixture_hg19[, human_cells]
mixture_mm10_mouseonly <- mixture_mm10[, mouse_cells]
dim(mixture_hg19_humanonly)[1] 33354 16163dim(mixture_mm10_mouseonly)[1] 28692 11338gene_ids_human <- rownames(mixture_hg19_humanonly)
gene_names_human <- make.unique(stringr::str_split_fixed(gene_ids_human, pattern = "_", n = 4)[, 4])
rownames(mixture_hg19_humanonly) <- gene_names_human
gene_ids_mouse <- rownames(mixture_mm10_mouseonly)
gene_names_mouse <- make.unique(stringr::str_split_fixed(gene_ids_mouse, pattern = "_", n = 4)[, 4])
rownames(mixture_mm10_mouseonly) <- gene_names_mousemixture_hg19_humanonly[1:5, 1:5]5 x 5 sparse Matrix of class "dgCMatrix"
Mixture1.Smart-seq2.p2_B1 Mixture1.Smart-seq2.p2_E12
TSPAN6 270 87
TNMD . .
DPM1 . 197
SCYL3 . .
C1orf112 213 99
Mixture1.Smart-seq2.p2_E7 Mixture1.Smart-seq2.p2_F4
TSPAN6 . 444
TNMD . .
DPM1 . 106
SCYL3 . 87
C1orf112 . 90
Mixture1.Smart-seq2.p2_G4
TSPAN6 288
TNMD .
DPM1 .
SCYL3 .
C1orf112 .mixture_mm10_mouseonly[1:5, 1:5]5 x 5 sparse Matrix of class "dgCMatrix"
Mixture1.Smart-seq2.p2_A4 Mixture1.Smart-seq2.p2_A7
Gnai3 181 115
Pbsn . .
Cdc45 . 100
H19 . 198
Scml2 . .
Mixture1.Smart-seq2.p1_A7 Mixture1.Smart-seq2.p2_B12
Gnai3 . .
Pbsn . .
Cdc45 . .
H19 . .
Scml2 . .
Mixture1.Smart-seq2.p2_B7
Gnai3 90
Pbsn .
Cdc45 61
H19 2256
Scml2 .mixture_hg19 <- CreateSeuratObject(counts = mixture_hg19_humanonly, project = "Ding_scBenchmark_mixturehg19", min.cells=1, min.features = 1)Warning: Feature names cannot have underscores ('_'), replacing with dashes
('-')mixture_hg19 <- AddMetaData(object = mixture_hg19, metadata = metadata[human_cells, ])
mixture_mm10 <- CreateSeuratObject(counts = mixture_mm10_mouseonly, project = "Ding_scBenchmark_mixturemm10", min.cells=1, min.features = 1)
mixture_mm10 <- AddMetaData(object = mixture_mm10, metadata = metadata[mouse_cells, ])umi_techs <- c("10x-Chromium-v2", "CEL-Seq2", "Drop-seq", "inDrops", "sci-RNA-seq")
clean_named_techs <- c("ChromiumV2", "CEL-seq2", "Drop-seq", "inDrops", "sci-RNA-seq")
names(clean_named_techs) <- umi_techs
clean_named_techs10x-Chromium-v2 CEL-Seq2 Drop-seq inDrops sci-RNA-seq
"ChromiumV2" "CEL-seq2" "Drop-seq" "inDrops" "sci-RNA-seq" `%notin%` <- Negate(`%in%`)
split_mixture_hg19 <- SplitObject(object = mixture_hg19, split.by = "technology")
names(split_mixture_hg19)[1] "Smart-seq2" "CEL-Seq2" "10x-Chromium-v2" "Drop-seq"
[5] "Seq-Well" "inDrops" "sci-RNA-seq" for (technology in names(split_mixture_hg19)) {
if (technology %notin% umi_techs) next
obj <- split_mixture_hg19[[technology]]
obj_split <- SplitObject(object = obj, split.by = "replicate")
clean_tech <- clean_named_techs[[technology]]
for (sampletype in names(obj_split)) {
seu_x <- obj_split[[sampletype]]
seu_x[["percent.mt"]] <- PercentageFeatureSet(seu_x, pattern = "^MT-")
saveRDS(
seu_x,
here::here("data/rds_raw", paste0("Ding-Human", sampletype, "__", clean_tech, ".rds"))
)
}
}
rm(split_mixture_hg19)
rm(mixture_hg19)split_mixture_mm10 <- SplitObject(object = mixture_mm10, split.by = "technology")
names(split_mixture_mm10)[1] "Smart-seq2" "CEL-Seq2" "10x-Chromium-v2" "Drop-seq"
[5] "Seq-Well" "inDrops" "sci-RNA-seq" for (technology in names(split_mixture_mm10)) {
if (technology %notin% umi_techs) next
obj <- split_mixture_mm10[[technology]]
obj_split <- SplitObject(object = obj, split.by = "replicate")
clean_tech <- clean_named_techs[[technology]]
for (sampletype in names(obj_split)) {
seu_x <- obj_split[[sampletype]]
seu_x[["percent.mt"]] <- PercentageFeatureSet(seu_x, pattern = "^mt-")
saveRDS(
seu_x,
here::here("data/rds_raw", paste0("Ding-Mouse", sampletype, "__", clean_tech, ".rds"))
)
}
}sessionInfo()R version 4.1.2 (2021-11-01)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 20.04.3 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.9.0
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.9.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] XML_3.99-0.8 httr_1.4.2 stringr_1.4.0 patchwork_1.1.1
[5] ggplot2_3.3.5 SeuratObject_4.0.4 Seurat_4.0.5 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] Rtsne_0.15 colorspace_2.0-2 deldir_1.0-6
[4] ellipsis_0.3.2 ggridges_0.5.3 rprojroot_2.0.2
[7] fs_1.5.2 spatstat.data_2.1-0 farver_2.1.0
[10] leiden_0.3.9 listenv_0.8.0 ggrepel_0.9.1
[13] fansi_0.5.0 codetools_0.2-18 splines_4.1.2
[16] knitr_1.36 polyclip_1.10-0 jsonlite_1.7.2
[19] ica_1.0-2 cluster_2.1.2 png_0.1-7
[22] uwot_0.1.11 shiny_1.7.1 sctransform_0.3.2.9008
[25] spatstat.sparse_2.0-0 compiler_4.1.2 assertthat_0.2.1
[28] Matrix_1.4-0 fastmap_1.1.0 lazyeval_0.2.2
[31] later_1.3.0 htmltools_0.5.2 tools_4.1.2
[34] igraph_1.2.9 gtable_0.3.0 glue_1.5.1
[37] RANN_2.6.1 reshape2_1.4.4 dplyr_1.0.7
[40] Rcpp_1.0.7 scattermore_0.7 jquerylib_0.1.4
[43] vctrs_0.3.8 nlme_3.1-152 lmtest_0.9-39
[46] xfun_0.28 globals_0.14.0 mime_0.12
[49] miniUI_0.1.1.1 lifecycle_1.0.1 irlba_2.3.5
[52] goftest_1.2-3 future_1.23.0 MASS_7.3-54
[55] zoo_1.8-9 scales_1.1.1 spatstat.core_2.3-2
[58] promises_1.2.0.1 spatstat.utils_2.3-0 parallel_4.1.2
[61] RColorBrewer_1.1-2 curl_4.3.2 yaml_2.2.1
[64] reticulate_1.22 pbapply_1.5-0 gridExtra_2.3
[67] sass_0.4.0 rpart_4.1-15 stringi_1.7.6
[70] highr_0.9 rlang_0.4.12 pkgconfig_2.0.3
[73] matrixStats_0.61.0 evaluate_0.14 lattice_0.20-45
[76] ROCR_1.0-11 purrr_0.3.4 tensor_1.5
[79] labeling_0.4.2 htmlwidgets_1.5.4 cowplot_1.1.1
[82] tidyselect_1.1.1 here_1.0.1 parallelly_1.29.0
[85] RcppAnnoy_0.0.19 plyr_1.8.6 magrittr_2.0.1
[88] R6_2.5.1 generics_0.1.1 DBI_1.1.1
[91] withr_2.4.3 mgcv_1.8-38 pillar_1.6.4
[94] whisker_0.4 fitdistrplus_1.1-6 survival_3.2-13
[97] abind_1.4-5 tibble_3.1.6 future.apply_1.8.1
[100] crayon_1.4.2 KernSmooth_2.23-20 utf8_1.2.2
[103] spatstat.geom_2.3-1 plotly_4.10.0 rmarkdown_2.11
[106] grid_4.1.2 data.table_1.14.2 git2r_0.29.0
[109] digest_0.6.29 xtable_1.8-4 tidyr_1.1.4
[112] httpuv_1.6.3 munsell_0.5.0 viridisLite_0.4.0
[115] bslib_0.3.1
sessionInfo()R version 4.1.2 (2021-11-01)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 20.04.3 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.9.0
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.9.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] XML_3.99-0.8 httr_1.4.2 stringr_1.4.0 patchwork_1.1.1
[5] ggplot2_3.3.5 SeuratObject_4.0.4 Seurat_4.0.5 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] Rtsne_0.15 colorspace_2.0-2 deldir_1.0-6
[4] ellipsis_0.3.2 ggridges_0.5.3 rprojroot_2.0.2
[7] fs_1.5.2 spatstat.data_2.1-0 farver_2.1.0
[10] leiden_0.3.9 listenv_0.8.0 ggrepel_0.9.1
[13] fansi_0.5.0 codetools_0.2-18 splines_4.1.2
[16] knitr_1.36 polyclip_1.10-0 jsonlite_1.7.2
[19] ica_1.0-2 cluster_2.1.2 png_0.1-7
[22] uwot_0.1.11 shiny_1.7.1 sctransform_0.3.2.9008
[25] spatstat.sparse_2.0-0 compiler_4.1.2 assertthat_0.2.1
[28] Matrix_1.4-0 fastmap_1.1.0 lazyeval_0.2.2
[31] later_1.3.0 htmltools_0.5.2 tools_4.1.2
[34] igraph_1.2.9 gtable_0.3.0 glue_1.5.1
[37] RANN_2.6.1 reshape2_1.4.4 dplyr_1.0.7
[40] Rcpp_1.0.7 scattermore_0.7 jquerylib_0.1.4
[43] vctrs_0.3.8 nlme_3.1-152 lmtest_0.9-39
[46] xfun_0.28 globals_0.14.0 mime_0.12
[49] miniUI_0.1.1.1 lifecycle_1.0.1 irlba_2.3.5
[52] goftest_1.2-3 future_1.23.0 MASS_7.3-54
[55] zoo_1.8-9 scales_1.1.1 spatstat.core_2.3-2
[58] promises_1.2.0.1 spatstat.utils_2.3-0 parallel_4.1.2
[61] RColorBrewer_1.1-2 curl_4.3.2 yaml_2.2.1
[64] reticulate_1.22 pbapply_1.5-0 gridExtra_2.3
[67] sass_0.4.0 rpart_4.1-15 stringi_1.7.6
[70] highr_0.9 rlang_0.4.12 pkgconfig_2.0.3
[73] matrixStats_0.61.0 evaluate_0.14 lattice_0.20-45
[76] ROCR_1.0-11 purrr_0.3.4 tensor_1.5
[79] labeling_0.4.2 htmlwidgets_1.5.4 cowplot_1.1.1
[82] tidyselect_1.1.1 here_1.0.1 parallelly_1.29.0
[85] RcppAnnoy_0.0.19 plyr_1.8.6 magrittr_2.0.1
[88] R6_2.5.1 generics_0.1.1 DBI_1.1.1
[91] withr_2.4.3 mgcv_1.8-38 pillar_1.6.4
[94] whisker_0.4 fitdistrplus_1.1-6 survival_3.2-13
[97] abind_1.4-5 tibble_3.1.6 future.apply_1.8.1
[100] crayon_1.4.2 KernSmooth_2.23-20 utf8_1.2.2
[103] spatstat.geom_2.3-1 plotly_4.10.0 rmarkdown_2.11
[106] grid_4.1.2 data.table_1.14.2 git2r_0.29.0
[109] digest_0.6.29 xtable_1.8-4 tidyr_1.1.4
[112] httpuv_1.6.3 munsell_0.5.0 viridisLite_0.4.0
[115] bslib_0.3.1