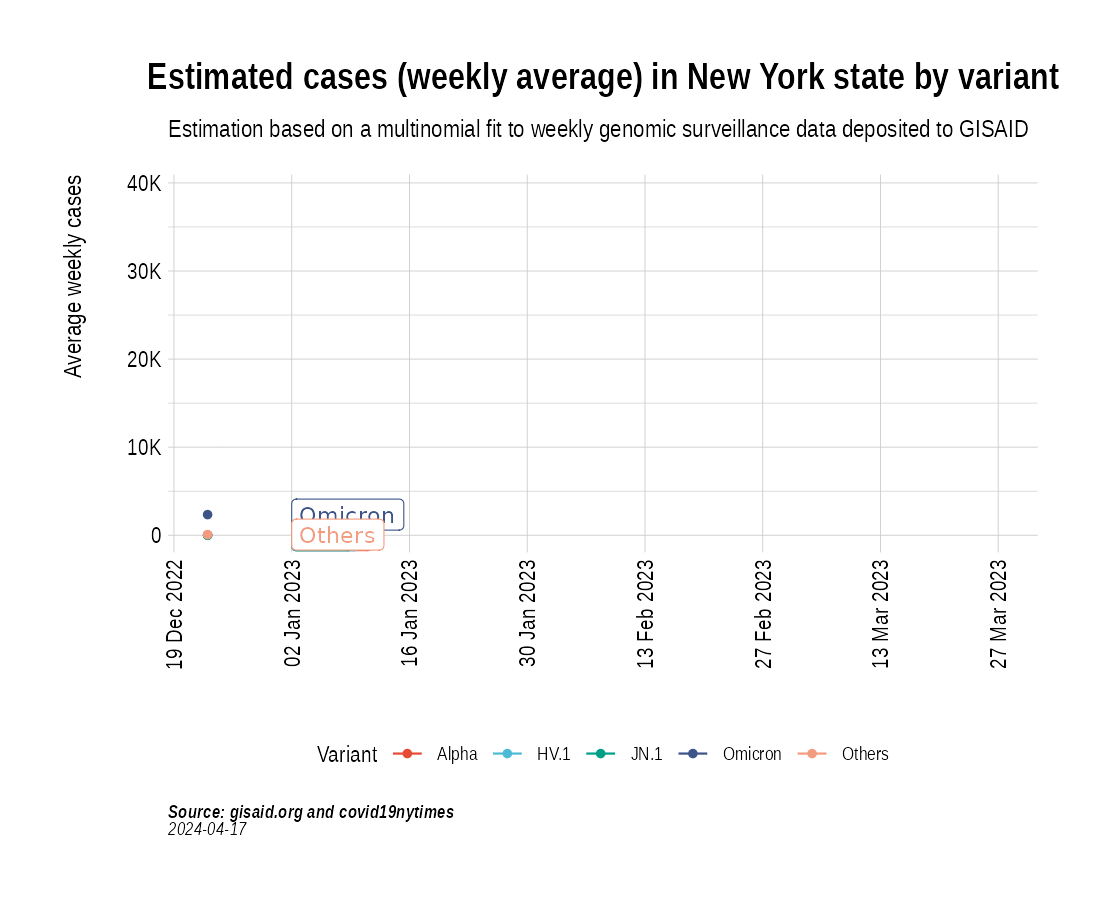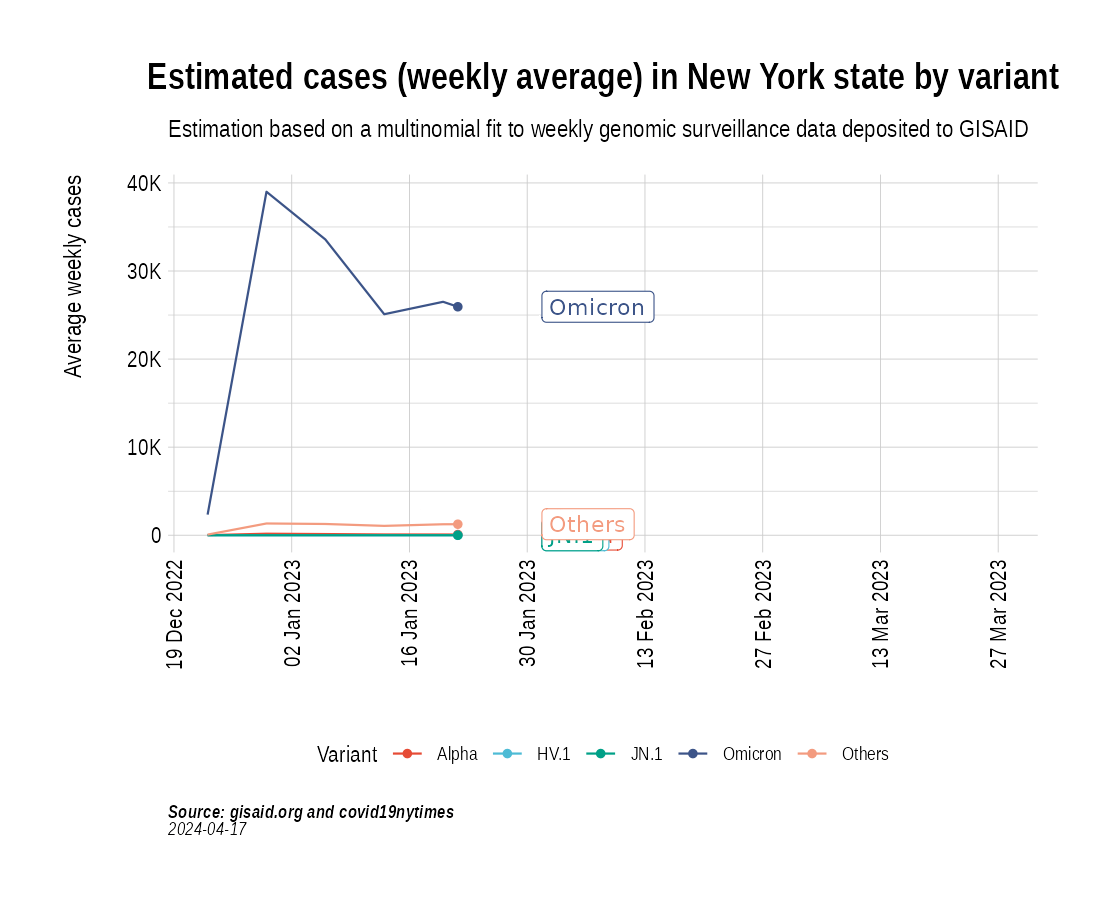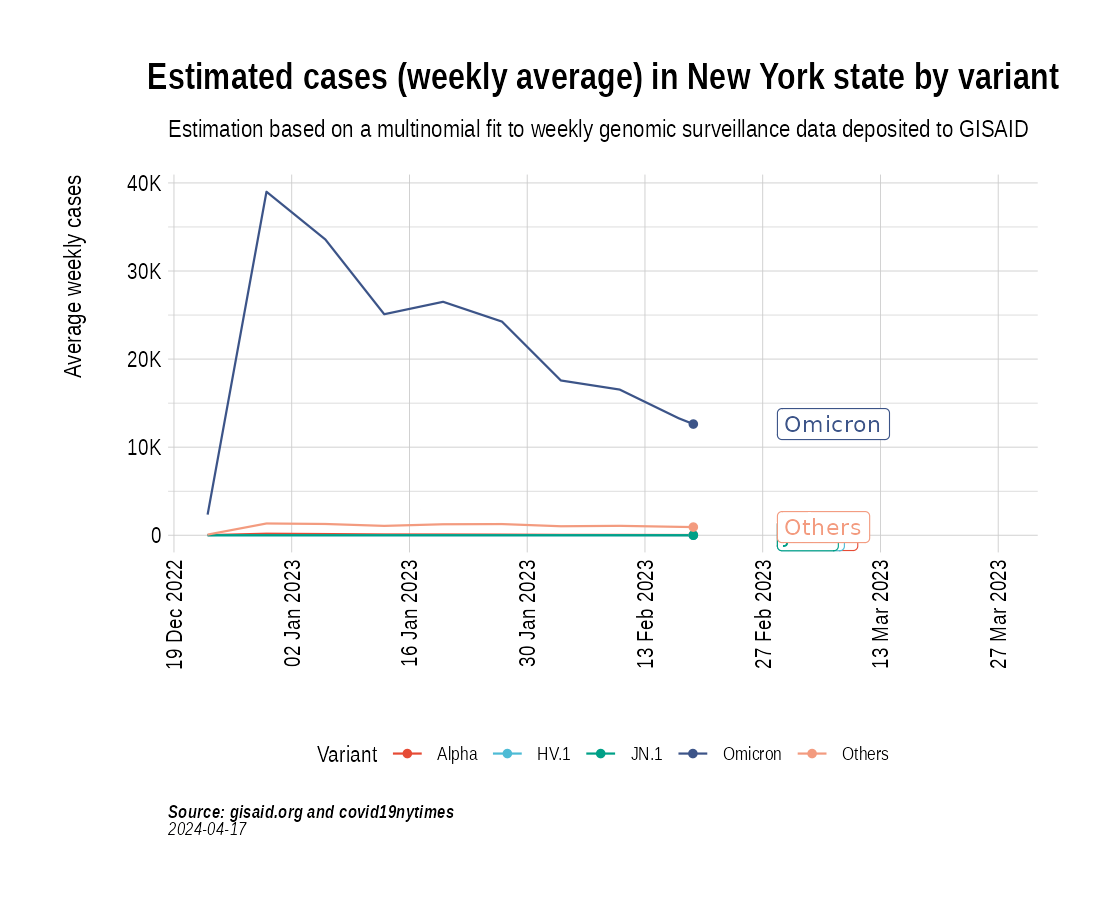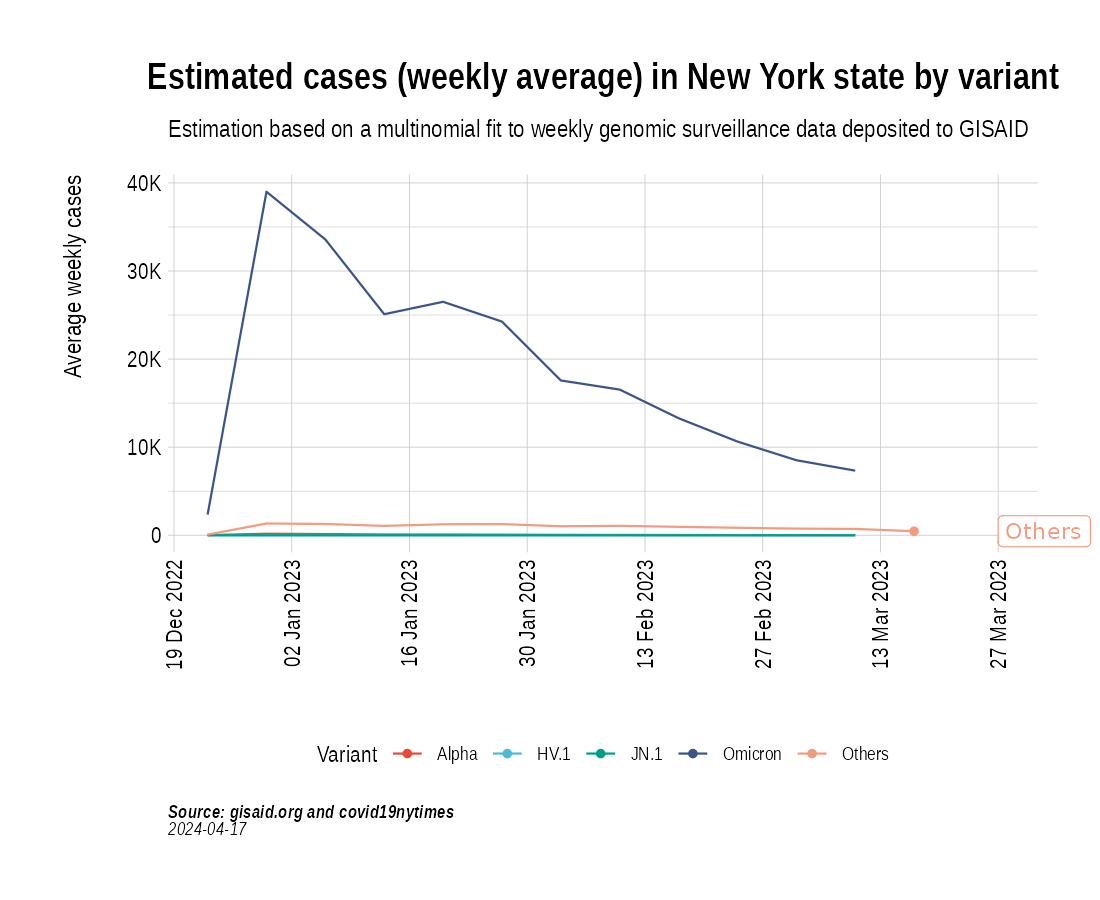
Animation of projected weekly cases - New York State
Source:vignettes/VariantAnimation-NewYork.Rmd
VariantAnimation-NewYork.Rmd
suppressPackageStartupMessages({
library(covmuller)
library(COVID19)
library(tidyverse)
})
theme_set(CovmullerTheme())Get variants data for NY state
gisaid_metadata <- qs::qread("~/data/epicov/metadata_tsv_2024_04_11.qs")
gisaid_usa <- gisaid_metadata %>%
filter(Country == "USA") %>%
filter(Host == "Human")
# format metadata
gisaid_usa <- FormatGISAIDMetadata(gisaid_usa)
gisaid_usa <- gisaid_usa %>%
arrange(State, MonthYearCollected) %>%
filter(pangolin_lineage != "Unknown")
gisaid_NY <- gisaid_usa %>% filter(State == "New York")
vocs <- GetVOCs()
custom_voc_mapping <- list(
`JN.1` = "JN.1",
`JN.1.*` = "JN.1",
`HV.1` = "HV.1",
`HV.1.*` = "HV.1",
`B.1` = "B.1",
`B.1.1.306` = "B.1",
`B.1.1.306.*` = "B.1",
`B.1.1.326` = "B.1",
`B.1.36.29` = "B.1",
`B.1.560` = "B.1",
`B.1.1` = "B.1",
`B.1.210` = "B.1",
`B.1.36.8` = "B.1",
`B.1.36` = "B.1",
`B.1.36.*` = "B.1"
)
gisaid_NY <- gisaid_NY %>% filter(pangolin_lineage != "None")
gisaid_NY <- CollapseLineageToVOCs(
variant_df = gisaid_NY,
vocs = vocs,
custom_voc_mapping = custom_voc_mapping,
summarize = FALSE
)Get weekly cases for New York
confirmed <- covid19(country = "USA", level = 2) %>%
filter(administrative_area_level_2 == "New York") %>%
select(date, confirmed) %>%
filter(!is.na(confirmed))
#> We have invested a lot of time and effort in creating COVID-19 Data
#> Hub, please cite the following when using it:
#>
#> Guidotti, E., Ardia, D., (2020), "COVID-19 Data Hub", Journal of Open
#> Source Software 5(51):2376, doi: 10.21105/joss.02376
#>
#> The implementation details and the latest version of the data are
#> described in:
#>
#> Guidotti, E., (2022), "A worldwide epidemiological database for
#> COVID-19 at fine-grained spatial resolution", Sci Data 9(1):112, doi:
#> 10.1038/s41597-022-01245-1
#> To print citations in BibTeX format use:
#> > print(citation('COVID19'), bibtex=TRUE)
#>
#> To hide this message use 'verbose = FALSE'.
confirmed$daily_cases <- c(confirmed$confirmed[1], diff(confirmed$confirmed))
confirmed$WeekYear <- tsibble::yearweek(confirmed$date)
confirmed$MonthYear <- GetMonthYear(confirmed$date)
confirmed_subset_dateweekwise_long <- confirmed %>%
group_by(WeekYear) %>%
summarise(n = ceiling(mean(daily_cases, na.rm = T))) %>%
arrange(WeekYear) %>%
rename(WeekYearCollected = WeekYear)
gisaid_NY_weekwise <- SummarizeVariantsWeekwise(gisaid_NY)Distribution of variants
state_month_counts <- SummarizeVariantsMonthwise(gisaid_NY)
state_month_counts$State <- "New York"
state_month_prevalence <- CountsToPrevalence(state_month_counts)
state_month_prevalence <- CollapseLineageToVOCs(
variant_df = state_month_prevalence,
vocs = vocs,
custom_voc_mapping = custom_voc_mapping, summarize = FALSE
)
p5 <- StackedBarPlotPrevalence(state_month_prevalence)
p5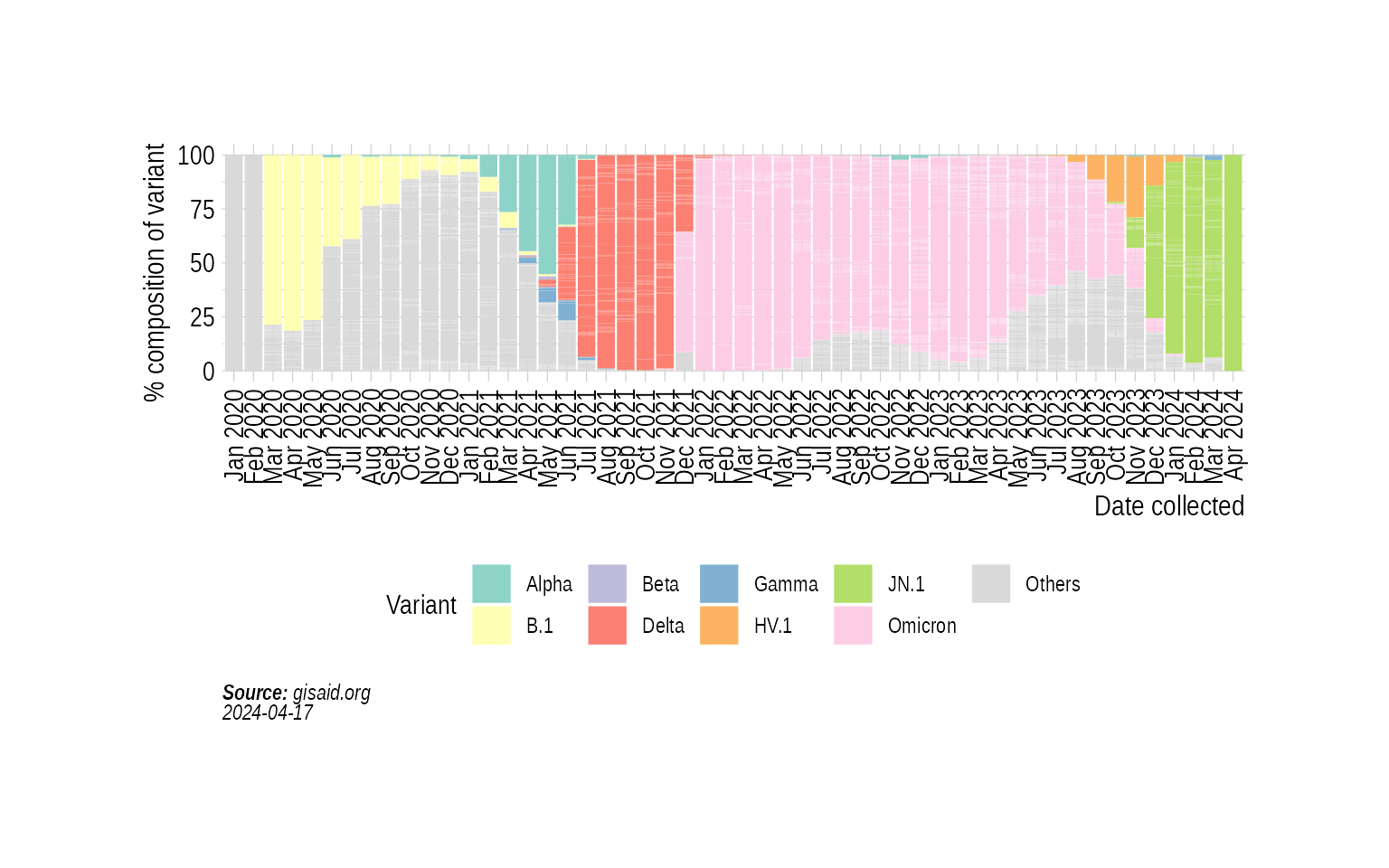
Project weekly cases to variant prevalence data from GISAID
voc_to_keep <- gisaid_NY_weekwise %>%
group_by(lineage_collapsed) %>%
summarise(n_sum = sum(n)) %>%
filter(n_sum > 50) %>%
pull(lineage_collapsed) %>%
unique()
gisaid_NY_weekwise <- gisaid_NY_weekwise %>% filter(lineage_collapsed %in% voc_to_keep)
newyork_cases_pred_prob_sel_long <- FitMultinomWeekly(gisaid_NY_weekwise, confirmed_subset_dateweekwise_long)
the_anim <- PlotVariantPrevalenceAnimated(newyork_cases_pred_prob_sel_long, title = "Estimated cases (weekly average) in New York state by variant", caption = "**Source: gisaid.org and covid19nytimes**<br>", date_breaks = "28 days")
gganimate::anim_save(filename = here::here("docs/articles/NY_animated.gif"), animation = the_anim)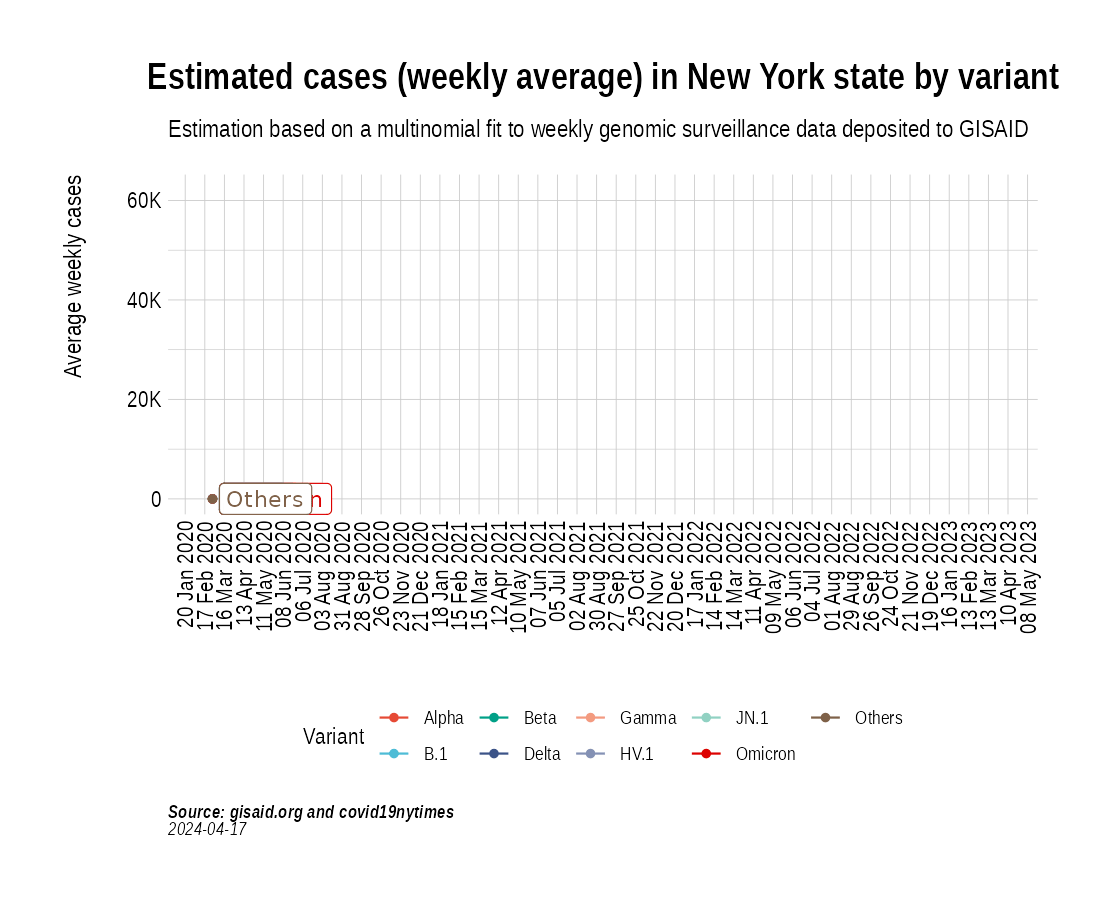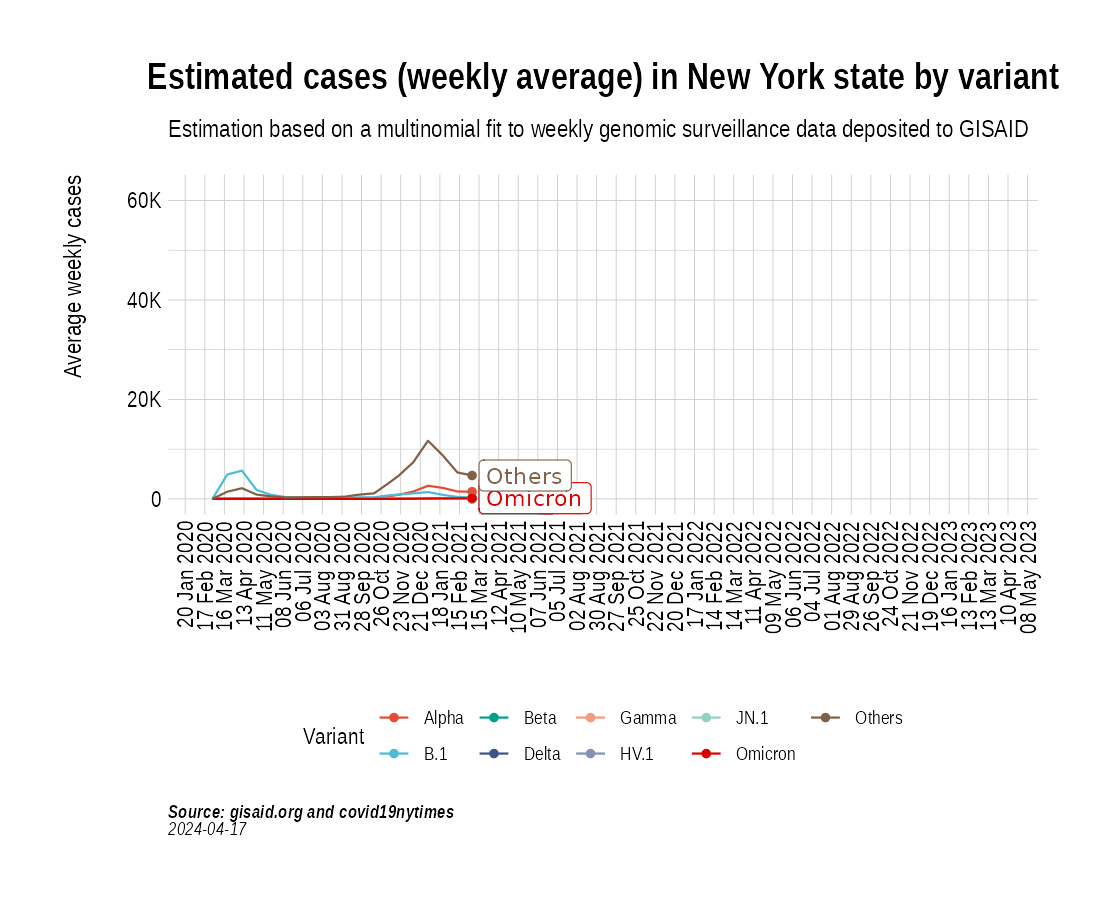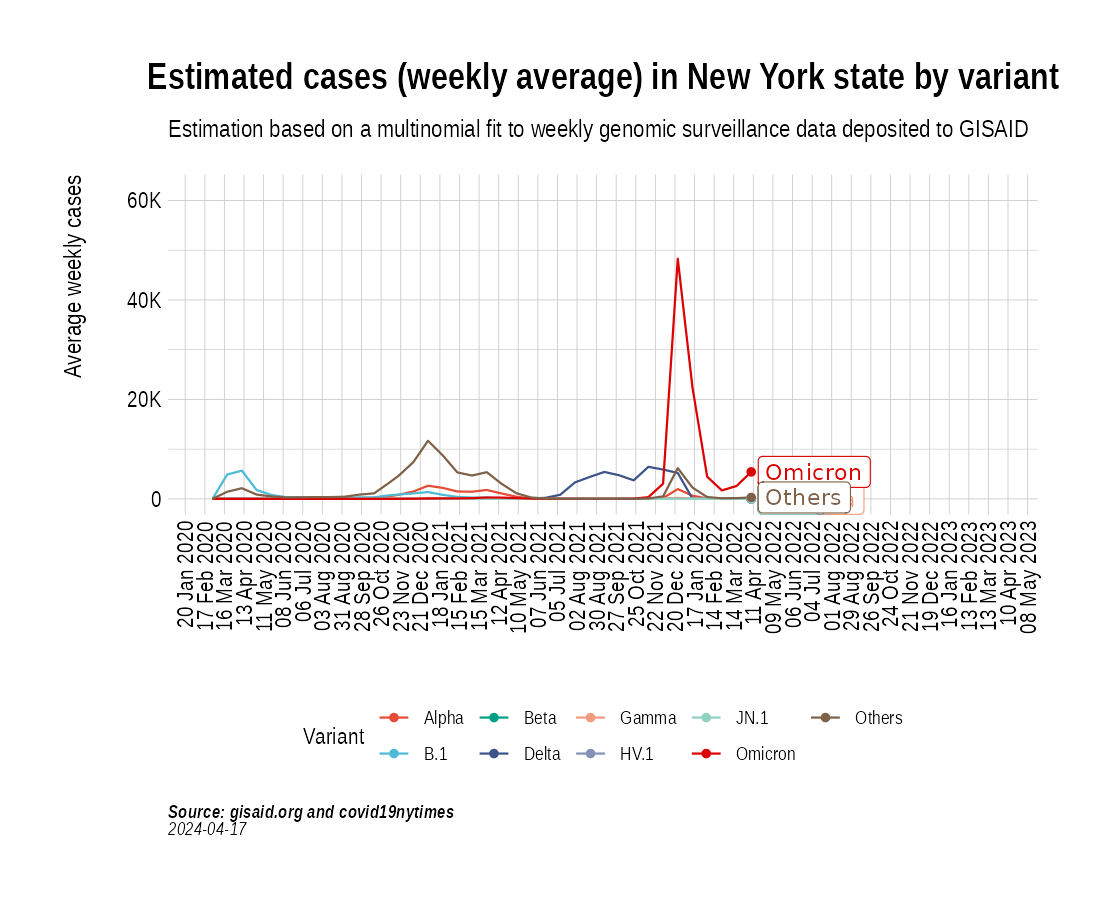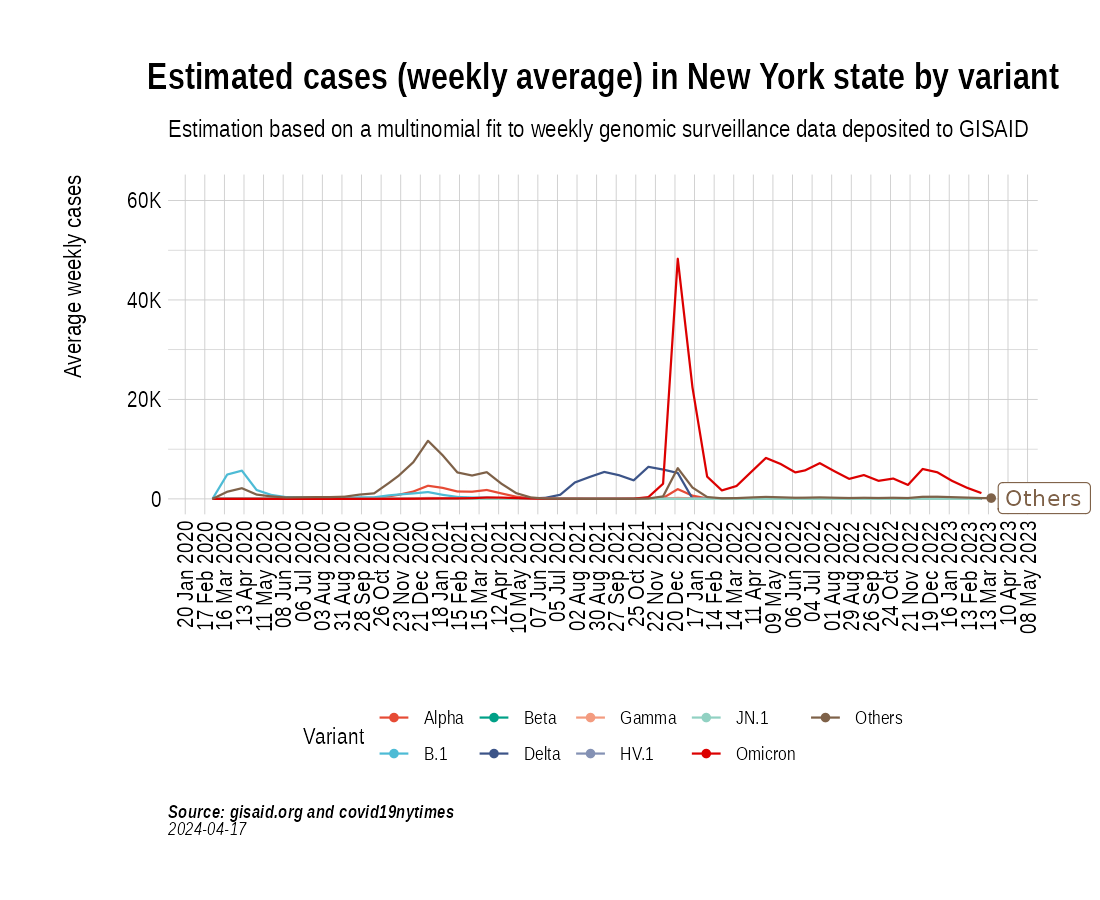
Look at cases after february, 2022 only:
confirmed$MonthYear <- GetMonthYear(confirmed$date)
confirmed_subset_dateweekwise_long <- confirmed %>%
filter(MonthYear > "December 2022") %>%
group_by(WeekYear) %>%
summarise(n = sum(daily_cases)) %>%
arrange(WeekYear) %>%
rename(WeekYearCollected = WeekYear)
gisaid_NY <- gisaid_NY %>% filter(MonthYearCollected > "December 2022")
gisaid_weekwise <- SummarizeVariantsWeekwise(gisaid_NY)
voc_to_keep <- gisaid_weekwise %>%
group_by(lineage_collapsed) %>%
summarise(n_sum = sum(n)) %>%
filter(n_sum > 50) %>%
pull(lineage_collapsed) %>%
unique()
gisaid_weekwise <- gisaid_weekwise %>% filter(lineage_collapsed %in% voc_to_keep)
cases_pred_prob_sel_long <- FitMultinomWeekly(gisaid_weekwise, confirmed_subset_dateweekwise_long)
the_anim <- PlotVariantPrevalenceAnimated(cases_pred_prob_sel_long, title = "Estimated cases (weekly average) in New York state by variant", caption = "**Source: gisaid.org and covid19nytimes**<br>")
gganimate::anim_save(filename = here::here("docs/articles/NY_animated_2023.gif"), animation = the_anim)