Quickstart¶
Most features in pysradb are accessible both from the command-line and
as a python package. pysradb usage on the two platforms will be
displayed by selecting the corresponding tab below.
Note
If you have any questions along the way, please head over to the Python API Usage or the Command Line for more information. You may also wish to refer to the API Documentation.
Notebooks¶
A Google Colaboratory version of most used commands are available in
this Colab
Notebook
. Colab runs Python 3.6 while pysradb requires Python 3.7+ and hence
the notebooks no longer run on Colab, but can be downloaded and run
locally.
The following notebooks document all the possible features of pysradb:
Metadata¶
pysradb makes it very easy to obtain metadata from SRA/EBI:
$ pysradb metadata SRP265425
from pysradb.sraweb import SRAweb
client = SRAweb()
df = client.metadata("SRP265425")
df
Output:
study_accession experiment_accession experiment_title experiment_desc organism_taxid organism_name library_name library_strategy library_source library_selection library_layout sample_accession sample_title instrument instrument_model instrument_model_desc total_spots total_size run_accession run_total_spots run_total_bases
SRP265425 SRX8434255 Ampliseq of SARS-CoV-2 Ampliseq of SARS-CoV-2 2697049 Severe acute respiratory syndrome coronavirus 2 63-2020-04-22 AMPLICON VIRAL RNA RT-PCR SINGLE SRS6745319 Ion Torrent S5 XL Ion Torrent S5 XL ION_TORRENT 1311358 83306910 SRR11886735 1311358 109594216
SRP265425 SRX8434254 Ampliseq of SARS-CoV-2 Ampliseq of SARS-CoV-2 2697049 Severe acute respiratory syndrome coronavirus 2 62-2020-04-22 AMPLICON VIRAL RNA RT-PCR SINGLE SRS6745320 Ion Torrent S5 XL Ion Torrent S5 XL ION_TORRENT 2614109 204278682 SRR11886736 2614109 262305651
SRP265425 SRX8434253 Ampliseq of SARS-CoV-2 Ampliseq of SARS-CoV-2 2697049 Severe acute respiratory syndrome coronavirus 2 61-2020-04-22 AMPLICON VIRAL RNA RT-PCR SINGLE SRS6745318 Ion Torrent S5 XL Ion Torrent S5 XL ION_TORRENT 2286312 183516004 SRR11886737 2286312 263304134
SRP265425 SRX8434252 Ampliseq of SARS-CoV-2 Ampliseq of SARS-CoV-2 2697049 Severe acute respiratory syndrome coronavirus 2 60-2020-04-22 AMPLICON VIRAL RNA RT-PCR SINGLE SRS6745317 Ion Torrent S5 XL Ion Torrent S5 XL ION_TORRENT 5202567 507524965 SRR11886738 5202567 781291588
SRP265425 SRX8434251 Ampliseq of SARS-CoV-2 Ampliseq of SARS-CoV-2 2697049 Severe acute respiratory syndrome coronavirus 2 38-2020-04-03 AMPLICON VIRAL RNA RT-PCR SINGLE SRS6745315 Ion Torrent S5 XL Ion Torrent S5 XL ION_TORRENT 3313960 356104406 SRR11886739 3313960 612430817
SRP265425 SRX8434250 Ampliseq of SARS-CoV-2 Ampliseq of SARS-CoV-2 2697049 Severe acute respiratory syndrome coronavirus 2 37-2020-04-03 AMPLICON VIRAL RNA RT-PCR SINGLE SRS6745316 Ion Torrent S5 XL Ion Torrent S5 XL ION_TORRENT 5155733 565882351 SRR11886740 5155733 954342917
SRP265425 SRX8434249 Ampliseq of SARS-CoV-2 Ampliseq of SARS-CoV-2 2697049 Severe acute respiratory syndrome coronavirus 2 36-2020-04-03 AMPLICON VIRAL RNA RT-PCR SINGLE SRS6745313 Ion Torrent S5 XL Ion Torrent S5 XL ION_TORRENT 1324589 175619046 SRR11886741 1324589 216531400
SRP265425 SRX8434248 Ampliseq of SARS-CoV-2 Ampliseq of SARS-CoV-2 2697049 Severe acute respiratory syndrome coronavirus 2 35-2020-04-03 AMPLICON VIRAL RNA RT-PCR SINGLE SRS6745314 Ion Torrent S5 XL Ion Torrent S5 XL ION_TORRENT 1639851 198973268 SRR11886742 1639851 245466005
SRP265425 SRX8434247 Ampliseq of SARS-CoV-2 Ampliseq of SARS-CoV-2 2697049 Severe acute respiratory syndrome coronavirus 2 68-2020-05-07 AMPLICON VIRAL RNA RT-PCR SINGLE SRS6745312 Ion Torrent S5 XL Ion Torrent S5 XL ION_TORRENT 3921389 210198580 SRR11886743 3921389 332935558
SRP265425 SRX8434246 Ampliseq of SARS-CoV-2 Ampliseq of SARS-CoV-2 2697049 Severe acute respiratory syndrome coronavirus 2 66-2020-04-22 AMPLICON VIRAL RNA RT-PCR SINGLE SRS6745311 Ion Torrent S5 XL Ion Torrent S5 XL ION_TORRENT 14295475 2150005008 SRR11886744 14295475 2967829315
SRP265425 SRX8434245 Ampliseq of SARS-CoV-2 Ampliseq of SARS-CoV-2 2697049 Severe acute respiratory syndrome coronavirus 2 65-2020-04-22 AMPLICON VIRAL RNA RT-PCR SINGLE SRS6745310 Ion Torrent S5 XL Ion Torrent S5 XL ION_TORRENT 5124692 294846140 SRR11886745 5124692 431819462
SRP265425 SRX8434244 Ampliseq of SARS-CoV-2 Ampliseq of SARS-CoV-2 2697049 Severe acute respiratory syndrome coronavirus 2 64-2020-04-22 AMPLICON VIRAL RNA RT-PCR SINGLE SRS6745309 Ion Torrent S5 XL Ion Torrent S5 XL ION_TORRENT 2986306 205666872 SRR11886746 2986306 275400959
SRP265425 SRX8434243 Ampliseq of SARS-CoV-2 Ampliseq of SARS-CoV-2 2697049 Severe acute respiratory syndrome coronavirus 2 34-2020-04-03 AMPLICON VIRAL RNA RT-PCR SINGLE SRS6745308 Ion Torrent S5 XL Ion Torrent S5 XL ION_TORRENT 1182690 59471336 SRR11886747 1182690 86350631
SRP265425 SRX8434242 Ampliseq of SARS-CoV-2 Ampliseq of SARS-CoV-2 2697049 Severe acute respiratory syndrome coronavirus 2 33-2020-04-03 AMPLICON VIRAL RNA RT-PCR SINGLE SRS6745307 Ion Torrent S5 XL Ion Torrent S5 XL ION_TORRENT 6031816 749323230 SRR11886748 6031816 928054297
Additionally to obtain locations of .fastq/.sra files and other
metadata:
pysradb makes it very easy to obtain metadata from SRA/EBI:
$ pysradb metadata SRP265425 --detailed
from pysradb.sraweb import SRAweb
client = SRAweb()
df = client.metadata("SRP265425", detailed=True)
df
Output:
run_accession study_accession experiment_accession experiment_title experiment_desc organism_taxid organism_name library_name library_strategy library_source library_selection library_layout sample_accession sample_title instrument instrument_model instrument_model_desc total_spots total_size run_total_spots run_total_bases run_alias sra_url_alt1 sra_url_alt2 sra_url experiment_alias isolate collected_by collection_date geo_loc_name host host_disease isolation_source lat_lon BioSampleModel sra_url_alt3 ena_fastq_http ena_fastq_http_1 ena_fastq_http_2 ena_fastq_ftp ena_fastq_ftp_1 ena_fastq_ftp_2
SRR11886735 SRP265425 SRX8434255 Ampliseq of SARS-CoV-2 Ampliseq of SARS-CoV-2 2697049 Severe acute respiratory syndrome coronavirus 2 63-2020-04-22 AMPLICON VIRAL RNA RT-PCR SINGLE SRS6745319 Ion Torrent S5 XL Ion Torrent S5 XL ION_TORRENT 1311358 83306910 1311358 109594216 IonXpress_063_R_2020_04_22_15_56_22_user_GCEID-S5-58-SARS_CoV2_SA4.bam gs://sra-pub-src-9/SRR11886735/Wuhan_Hu_1_NC_045512_21500_and_subgenomics_SA4.fasta.1 https://sra-pub-sars-cov2.s3.amazonaws.com/sra-src/SRR11886735/Wuhan_Hu_1_NC_045512_21500_and_subgenomics_SA4.fasta.1 https://sra-download.ncbi.nlm.nih.gov/traces/sra0/SRR/011608/SRR11886735 GC-20 NA 02-Apr-2020 Australia: Victoria Homo sapiens COVID-19 swab NA Pathogen.cl http://ftp.sra.ebi.ac.uk/vol1/fastq/SRR118/035/SRR11886735/SRR11886735.fastq.gz era-fasp@fasp.sra.ebi.ac.uk:vol1/fastq/SRR118/035/SRR11886735/SRR11886735.fastq.gz
SRR11886736 SRP265425 SRX8434254 Ampliseq of SARS-CoV-2 Ampliseq of SARS-CoV-2 2697049 Severe acute respiratory syndrome coronavirus 2 62-2020-04-22 AMPLICON VIRAL RNA RT-PCR SINGLE SRS6745320 Ion Torrent S5 XL Ion Torrent S5 XL ION_TORRENT 2614109 204278682 2614109 262305651 IonXpress_062_R_2020_04_22_15_56_22_user_GCEID-S5-58-SARS_CoV2_SA4.bam gs://sra-pub-src-16/SRR11886736/Wuhan_Hu_1_NC_045512_21500_and_subgenomics_SA4.fasta.1 https://sra-download.ncbi.nlm.nih.gov/traces/sra46/SRZ/011886/SRR11886736/Wuhan_Hu_1_NC_045512_21500_and_subgenomics_SA4.fasta https://sra-download.ncbi.nlm.nih.gov/traces/sra50/SRR/011608/SRR11886736 GC-51 NA 14-Apr-2020 Australia: Victoria Homo sapiens COVID-19 swab NA Pathogen.cl https://sra-pub-sars-cov2.s3.amazonaws.com/sra-src/SRR11886736/Wuhan_Hu_1_NC_045512_21500_and_subgenomics_SA4.fasta.1 http://ftp.sra.ebi.ac.uk/vol1/fastq/SRR118/036/SRR11886736/SRR11886736.fastq.gz era-fasp@fasp.sra.ebi.ac.uk:vol1/fastq/SRR118/036/SRR11886736/SRR11886736.fastq.gz
SRR11886737 SRP265425 SRX8434253 Ampliseq of SARS-CoV-2 Ampliseq of SARS-CoV-2 2697049 Severe acute respiratory syndrome coronavirus 2 61-2020-04-22 AMPLICON VIRAL RNA RT-PCR SINGLE SRS6745318 Ion Torrent S5 XL Ion Torrent S5 XL ION_TORRENT 2286312 183516004 2286312 263304134 IonXpress_061_R_2020_04_22_15_56_22_user_GCEID-S5-58-SARS_CoV2_SA4.bam gs://sra-pub-src-16/SRR11886737/Wuhan_Hu_1_NC_045512_21500_and_subgenomics_SA4.fasta.1 https://sra-download.ncbi.nlm.nih.gov/traces/sra29/SRZ/011886/SRR11886737/Wuhan_Hu_1_NC_045512_21500_and_subgenomics_SA4.fasta https://sra-download.ncbi.nlm.nih.gov/traces/sra17/SRR/011608/SRR11886737 GC-24 NA 07-Apr-2020 Australia: Victoria Homo sapiens COVID-19 swab NA Pathogen.cl https://sra-pub-sars-cov2.s3.amazonaws.com/sra-src/SRR11886737/Wuhan_Hu_1_NC_045512_21500_and_subgenomics_SA4.fasta.1 http://ftp.sra.ebi.ac.uk/vol1/fastq/SRR118/037/SRR11886737/SRR11886737.fastq.gz era-fasp@fasp.sra.ebi.ac.uk:vol1/fastq/SRR118/037/SRR11886737/SRR11886737.fastq.gz
SRR11886738 SRP265425 SRX8434252 Ampliseq of SARS-CoV-2 Ampliseq of SARS-CoV-2 2697049 Severe acute respiratory syndrome coronavirus 2 60-2020-04-22 AMPLICON VIRAL RNA RT-PCR SINGLE SRS6745317 Ion Torrent S5 XL Ion Torrent S5 XL ION_TORRENT 5202567 507524965 5202567 781291588 IonXpress_060_R_2020_04_22_15_56_22_user_GCEID-S5-58-SARS_CoV2_SA4.bam gs://sra-pub-src-15/SRR11886738/IonXpress_060_R_2020_04_22_15_56_22_user_GCEID_S5_58_SARS_CoV2_SA4.bam.1 https://sra-download.ncbi.nlm.nih.gov/traces/sra69/SRZ/011886/SRR11886738/IonXpress_060_R_2020_04_22_15_56_22_user_GCEID_S5_58_SARS_CoV2_SA4.bam https://sra-download.ncbi.nlm.nih.gov/traces/sra77/SRR/011608/SRR11886738 GC-23 NA 08-Apr-2020 Australia: Victoria Homo sapiens COVID-19 swab NA Pathogen.cl https://sra-pub-sars-cov2.s3.amazonaws.com/sra-src/SRR11886738/IonXpress_060_R_2020_04_22_15_56_22_user_GCEID_S5_58_SARS_CoV2_SA4.bam.1 http://ftp.sra.ebi.ac.uk/vol1/fastq/SRR118/038/SRR11886738/SRR11886738.fastq.gz era-fasp@fasp.sra.ebi.ac.uk:vol1/fastq/SRR118/038/SRR11886738/SRR11886738.fastq.gz
SRR11886739 SRP265425 SRX8434251 Ampliseq of SARS-CoV-2 Ampliseq of SARS-CoV-2 2697049 Severe acute respiratory syndrome coronavirus 2 38-2020-04-03 AMPLICON VIRAL RNA RT-PCR SINGLE SRS6745315 Ion Torrent S5 XL Ion Torrent S5 XL ION_TORRENT 3313960 356104406 3313960 612430817 IonXpress_038_R_2020_04_03_10_09_05_user_GCEID-S5-55-SARS_CoV2_SA4.bam gs://sra-pub-src-13/SRR11886739/Wuhan_Hu_1_NC_045512_21500_and_subgenomics_SA4.fasta.1 https://sra-pub-sars-cov2.s3.amazonaws.com/sra-src/SRR11886739/Wuhan_Hu_1_NC_045512_21500_and_subgenomics_SA4.fasta.1 https://sra-download.ncbi.nlm.nih.gov/traces/sra24/SRR/011608/SRR11886739 GC-11b NA 24-Mar-2020 Australia: Victoria Homo sapiens COVID-19 swab NA Pathogen.cl http://ftp.sra.ebi.ac.uk/vol1/fastq/SRR118/039/SRR11886739/SRR11886739.fastq.gz era-fasp@fasp.sra.ebi.ac.uk:vol1/fastq/SRR118/039/SRR11886739/SRR11886739.fastq.gz
SRR11886740 SRP265425 SRX8434250 Ampliseq of SARS-CoV-2 Ampliseq of SARS-CoV-2 2697049 Severe acute respiratory syndrome coronavirus 2 37-2020-04-03 AMPLICON VIRAL RNA RT-PCR SINGLE SRS6745316 Ion Torrent S5 XL Ion Torrent S5 XL ION_TORRENT 5155733 565882351 5155733 954342917 IonXpress_037_R_2020_04_03_10_09_05_user_GCEID-S5-55-SARS_CoV2_SA4.bam gs://sra-pub-src-5/SRR11886740/IonXpress_037_R_2020_04_03_10_09_05_user_GCEID_S5_55_SARS_CoV2_SA4.bam.1 https://sra-pub-sars-cov2.s3.amazonaws.com/sra-src/SRR11886740/IonXpress_037_R_2020_04_03_10_09_05_user_GCEID_S5_55_SARS_CoV2_SA4.bam.1 https://sra-download.ncbi.nlm.nih.gov/traces/sra13/SRR/011608/SRR11886740 GC-14b NA 28-Mar-2020 Australia: Victoria Homo sapiens COVID-19 swab NA Pathogen.cl http://ftp.sra.ebi.ac.uk/vol1/fastq/SRR118/040/SRR11886740/SRR11886740.fastq.gz era-fasp@fasp.sra.ebi.ac.uk:vol1/fastq/SRR118/040/SRR11886740/SRR11886740.fastq.gz
SRR11886741 SRP265425 SRX8434249 Ampliseq of SARS-CoV-2 Ampliseq of SARS-CoV-2 2697049 Severe acute respiratory syndrome coronavirus 2 36-2020-04-03 AMPLICON VIRAL RNA RT-PCR SINGLE SRS6745313 Ion Torrent S5 XL Ion Torrent S5 XL ION_TORRENT 1324589 175619046 1324589 216531400 IonXpress_036_R_2020_04_03_10_09_05_user_GCEID-S5-55-SARS_CoV2_SA4.bam gs://sra-pub-src-11/SRR11886741/Wuhan_Hu_1_NC_045512_21500_and_subgenomics_SA4.fasta.1 https://sra-pub-sars-cov2.s3.amazonaws.com/sra-src/SRR11886741/Wuhan_Hu_1_NC_045512_21500_and_subgenomics_SA4.fasta.1 https://sra-download.ncbi.nlm.nih.gov/traces/sra57/SRR/011608/SRR11886741 GC-12 NA 24-Mar-2020 Australia: Victoria Homo sapiens COVID-19 swab NA Pathogen.cl http://ftp.sra.ebi.ac.uk/vol1/fastq/SRR118/041/SRR11886741/SRR11886741.fastq.gz era-fasp@fasp.sra.ebi.ac.uk:vol1/fastq/SRR118/041/SRR11886741/SRR11886741.fastq.gz
SRR11886742 SRP265425 SRX8434248 Ampliseq of SARS-CoV-2 Ampliseq of SARS-CoV-2 2697049 Severe acute respiratory syndrome coronavirus 2 35-2020-04-03 AMPLICON VIRAL RNA RT-PCR SINGLE SRS6745314 Ion Torrent S5 XL Ion Torrent S5 XL ION_TORRENT 1639851 198973268 1639851 245466005 IonXpress_035_R_2020_04_03_10_09_05_user_GCEID-S5-55-SARS_CoV2_SA4.bam gs://sra-pub-src-11/SRR11886742/Wuhan_Hu_1_NC_045512_21500_and_subgenomics_SA4.fasta.1 https://sra-pub-sars-cov2.s3.amazonaws.com/sra-src/SRR11886742/Wuhan_Hu_1_NC_045512_21500_and_subgenomics_SA4.fasta.1 https://sra-download.ncbi.nlm.nih.gov/traces/sra69/SRR/011608/SRR11886742 GC-13 NA 23-Mar-2020 Australia: Victoria Homo sapiens COVID-19 swab NA Pathogen.cl http://ftp.sra.ebi.ac.uk/vol1/fastq/SRR118/042/SRR11886742/SRR11886742.fastq.gz era-fasp@fasp.sra.ebi.ac.uk:vol1/fastq/SRR118/042/SRR11886742/SRR11886742.fastq.gz
SRR11886743 SRP265425 SRX8434247 Ampliseq of SARS-CoV-2 Ampliseq of SARS-CoV-2 2697049 Severe acute respiratory syndrome coronavirus 2 68-2020-05-07 AMPLICON VIRAL RNA RT-PCR SINGLE SRS6745312 Ion Torrent S5 XL Ion Torrent S5 XL ION_TORRENT 3921389 210198580 3921389 332935558 IonXpress_068_R_2020_05_07_11_47_51_user_GCEID-S5-60-SARS_CoV2_SA4.bam gs://sra-pub-src-17/SRR11886743/Wuhan_Hu_1_NC_045512_21500_and_subgenomics_SA4.fasta.1 https://sra-download.ncbi.nlm.nih.gov/traces/sra64/SRZ/011886/SRR11886743/Wuhan_Hu_1_NC_045512_21500_and_subgenomics_SA4.fasta https://sra-download.ncbi.nlm.nih.gov/traces/sra54/SRR/011608/SRR11886743 GC-55 NA 24-Apr-2020 Australia: Victoria Homo sapiens COVID-19 swab NA Pathogen.cl https://sra-pub-sars-cov2.s3.amazonaws.com/sra-src/SRR11886743/Wuhan_Hu_1_NC_045512_21500_and_subgenomics_SA4.fasta.1 http://ftp.sra.ebi.ac.uk/vol1/fastq/SRR118/043/SRR11886743/SRR11886743.fastq.gz era-fasp@fasp.sra.ebi.ac.uk:vol1/fastq/SRR118/043/SRR11886743/SRR11886743.fastq.gz
SRR11886744 SRP265425 SRX8434246 Ampliseq of SARS-CoV-2 Ampliseq of SARS-CoV-2 2697049 Severe acute respiratory syndrome coronavirus 2 66-2020-04-22 AMPLICON VIRAL RNA RT-PCR SINGLE SRS6745311 Ion Torrent S5 XL Ion Torrent S5 XL ION_TORRENT 14295475 2150005008 14295475 2967829315 IonXpress_066_R_2020_04_22_11_10_56_user_GCEID-S5-57-SARS_CoV2_SA4.fastq gs://sra-pub-src-11/SRR11886744/IonXpress_066_R_2020_04_22_11_10_56_user_GCEID-S5-57-SARS_CoV2_SA4.fastq.1 https://sra-pub-sars-cov2.s3.amazonaws.com/sra-src/SRR11886744/IonXpress_066_R_2020_04_22_11_10_56_user_GCEID-S5-57-SARS_CoV2_SA4.fastq.1 https://sra-download.ncbi.nlm.nih.gov/traces/sra20/SRR/011608/SRR11886744 GC-26 NA 07-Mar-2020 Australia: Victoria Homo sapiens COVID-19 swab NA Pathogen.cl http://ftp.sra.ebi.ac.uk/vol1/fastq/SRR118/044/SRR11886744/SRR11886744.fastq.gz era-fasp@fasp.sra.ebi.ac.uk:vol1/fastq/SRR118/044/SRR11886744/SRR11886744.fastq.gz
SRR11886745 SRP265425 SRX8434245 Ampliseq of SARS-CoV-2 Ampliseq of SARS-CoV-2 2697049 Severe acute respiratory syndrome coronavirus 2 65-2020-04-22 AMPLICON VIRAL RNA RT-PCR SINGLE SRS6745310 Ion Torrent S5 XL Ion Torrent S5 XL ION_TORRENT 5124692 294846140 5124692 431819462 IonXpress_065_R_2020_04_22_11_10_56_user_GCEID-S5-57-SARS_CoV2_SA4.bam gs://sra-pub-src-16/SRR11886745/Wuhan_Hu_1_NC_045512_21500_and_subgenomics_SA4.fasta.1 https://sra-download.ncbi.nlm.nih.gov/traces/sra69/SRZ/011886/SRR11886745/Wuhan_Hu_1_NC_045512_21500_and_subgenomics_SA4.fasta https://sra-download.ncbi.nlm.nih.gov/traces/sra19/SRR/011608/SRR11886745 GC-25 NA 10-Apr-2020 Australia: Victoria Homo sapiens COVID-19 swab NA Pathogen.cl https://sra-pub-sars-cov2.s3.amazonaws.com/sra-src/SRR11886745/Wuhan_Hu_1_NC_045512_21500_and_subgenomics_SA4.fasta.1 http://ftp.sra.ebi.ac.uk/vol1/fastq/SRR118/045/SRR11886745/SRR11886745.fastq.gz era-fasp@fasp.sra.ebi.ac.uk:vol1/fastq/SRR118/045/SRR11886745/SRR11886745.fastq.gz
SRR11886746 SRP265425 SRX8434244 Ampliseq of SARS-CoV-2 Ampliseq of SARS-CoV-2 2697049 Severe acute respiratory syndrome coronavirus 2 64-2020-04-22 AMPLICON VIRAL RNA RT-PCR SINGLE SRS6745309 Ion Torrent S5 XL Ion Torrent S5 XL ION_TORRENT 2986306 205666872 2986306 275400959 IonXpress_064_R_2020_04_22_15_56_22_user_GCEID-S5-58-SARS_CoV2_SA4.bam gs://sra-pub-src-17/SRR11886746/Wuhan_Hu_1_NC_045512_21500_and_subgenomics_SA4.fasta.1 https://sra-download.ncbi.nlm.nih.gov/traces/sra59/SRZ/011886/SRR11886746/Wuhan_Hu_1_NC_045512_21500_and_subgenomics_SA4.fasta https://sra-download.ncbi.nlm.nih.gov/traces/sra47/SRR/011608/SRR11886746 GC-21 NA 03-Apr-2020 Australia: Victoria Homo sapiens COVID-19 swab NA Pathogen.cl https://sra-pub-sars-cov2.s3.amazonaws.com/sra-src/SRR11886746/Wuhan_Hu_1_NC_045512_21500_and_subgenomics_SA4.fasta.1 http://ftp.sra.ebi.ac.uk/vol1/fastq/SRR118/046/SRR11886746/SRR11886746.fastq.gz era-fasp@fasp.sra.ebi.ac.uk:vol1/fastq/SRR118/046/SRR11886746/SRR11886746.fastq.gz
SRR11886747 SRP265425 SRX8434243 Ampliseq of SARS-CoV-2 Ampliseq of SARS-CoV-2 2697049 Severe acute respiratory syndrome coronavirus 2 34-2020-04-03 AMPLICON VIRAL RNA RT-PCR SINGLE SRS6745308 Ion Torrent S5 XL Ion Torrent S5 XL ION_TORRENT 1182690 59471336 1182690 86350631 IonXpress_034_R_2020_04_03_10_09_05_user_GCEID-S5-55-SARS_CoV2_SA4.bam gs://sra-pub-src-16/SRR11886747/Wuhan_Hu_1_NC_045512_21500_and_subgenomics_SA4.fasta.1 https://sra-download.ncbi.nlm.nih.gov/traces/sra77/SRZ/011886/SRR11886747/Wuhan_Hu_1_NC_045512_21500_and_subgenomics_SA4.fasta https://sra-download.ncbi.nlm.nih.gov/traces/sra13/SRR/011608/SRR11886747 GC-11a NA 24-Mar-2020 Australia: Victoria Homo sapiens COVID-19 swab NA Pathogen.cl https://sra-pub-sars-cov2.s3.amazonaws.com/sra-src/SRR11886747/Wuhan_Hu_1_NC_045512_21500_and_subgenomics_SA4.fasta.1 http://ftp.sra.ebi.ac.uk/vol1/fastq/SRR118/047/SRR11886747/SRR11886747.fastq.gz era-fasp@fasp.sra.ebi.ac.uk:vol1/fastq/SRR118/047/SRR11886747/SRR11886747.fastq.gz
SRR11886748 SRP265425 SRX8434242 Ampliseq of SARS-CoV-2 Ampliseq of SARS-CoV-2 2697049 Severe acute respiratory syndrome coronavirus 2 33-2020-04-03 AMPLICON VIRAL RNA RT-PCR SINGLE SRS6745307 Ion Torrent S5 XL Ion Torrent S5 XL ION_TORRENT 6031816 749323230 6031816 928054297 IonXpress_033_R_2020_04_03_10_09_05_user_GCEID-S5-55-SARS_CoV2_SA4.bam gs://sra-pub-src-15/SRR11886748/IonXpress_033_R_2020_04_03_10_09_05_user_GCEID_S5_55_SARS_CoV2_SA4.bam.1 https://sra-download.ncbi.nlm.nih.gov/traces/sra43/SRZ/011886/SRR11886748/IonXpress_033_R_2020_04_03_10_09_05_user_GCEID_S5_55_SARS_CoV2_SA4.bam https://sra-download.ncbi.nlm.nih.gov/traces/sra66/SRR/011608/SRR11886748 GC-14a NA 28-Mar-2020 Australia: Victoria Homo sapiens COVID-19 swab NA Pathogen.cl https://sra-pub-sars-cov2.s3.amazonaws.com/sra-src/SRR11886748/IonXpress_033_R_2020_04_03_10_09_05_user_GCEID_S5_55_SARS_CoV2_SA4.bam.1 http://ftp.sra.ebi.ac.uk/vol1/fastq/SRR118/048/SRR11886748/SRR11886748.fastq.gz era-fasp@fasp.sra.ebi.ac.uk:vol1/fastq/SRR118/048/SRR11886748/SRR11886748.fastq.gz
Converting between accession numbers¶
pysradb provides a suite of commands for interoperability between
conversion numbers.
Convert SRP to SRX¶
$ pysradb srp-to-srx SRP098789
from pysradb.sraweb import SRAweb
client = SRAweb()
df = client.srp-to-srx("SRP098789")
df
Output:
study_accession experiment_accession experiment_title experiment_desc organism_taxid organism_name library_strategy library_source library_selection sample_accession sample_title instrument total_spots total_size run_accession run_total_spots run_total_bases study_accesssion
SRP098789 SRX2536428 GSM2476022: vehicle, 60 min, rep 5-mRNAseq; Homo sapiens; RNA-Seq GSM2476022: vehicle, 60 min, rep 5-mRNAseq; Homo sapiens; RNA-Seq 9606 Homo sapiens RNA-Seq TRANSCRIPTOMIC cDNA SRS1956378 Illumina HiSeq 2500 69422931 1545681856 SRR5227313 69422931 3540569481 SRP098789
SRP098789 SRX2536427 GSM2476021: PF-06446846, 60 min, rep 5 -mRNA-seq; Homo sapiens; OTHER GSM2476021: PF-06446846, 60 min, rep 5 -mRNA-seq; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956377 Illumina HiSeq 2500 58065134 1302369810 SRR5227312 58065134 2961321834 SRP098789
SRP098789 SRX2536426 GSM2476020: vehicle, 60 min, rep 4-mRNAseq; Homo sapiens; RNA-Seq GSM2476020: vehicle, 60 min, rep 4-mRNAseq; Homo sapiens; RNA-Seq 9606 Homo sapiens RNA-Seq TRANSCRIPTOMIC cDNA SRS1956376 Illumina HiSeq 2500 63720205 1416818619 SRR5227311 63720205 3249730455 SRP098789
SRP098789 SRX2536425 GSM2476019: PF-06446846, 60 min, rep 4 -mRNA-seq; Homo sapiens; OTHER GSM2476019: PF-06446846, 60 min, rep 4 -mRNA-seq; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956375 Illumina HiSeq 2500 66363585 1482728577 SRR5227310 66363585 3384542835 SRP098789
SRP098789 SRX2536424 GSM2476018: vehicle, 60 min, rep 5-Ribo-seq; Homo sapiens; RNA-Seq GSM2476018: vehicle, 60 min, rep 5-Ribo-seq; Homo sapiens; RNA-Seq 9606 Homo sapiens RNA-Seq TRANSCRIPTOMIC cDNA SRS1956374 Illumina HiSeq 2500 40062613 904488287 SRR5227309 40062613 2043193263 SRP098789
SRP098789 SRX2536423 GSM2476017: 1.5 ?M PF-067446846, 60 min, rep 5 -riboseq; Homo sapiens; OTHER GSM2476017: 1.5 ?M PF-067446846, 60 min, rep 5 -riboseq; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956373 Illumina HiSeq 2500 65591217 1499668100 SRR5227308 65591217 3345152067 SRP098789
SRP098789 SRX2536422 GSM2476016: Vehicle, 60 min, rep 4-ribo-seq; Homo sapiens; RNA-Seq GSM2476016: Vehicle, 60 min, rep 4-ribo-seq; Homo sapiens; RNA-Seq 9606 Homo sapiens RNA-Seq TRANSCRIPTOMIC cDNA SRS1956372 Illumina HiSeq 2500 66480991 1564636133 SRR5227307 66480991 3390530541 SRP098789
SRP098789 SRX2536421 GSM2476015: 1.5 ?M PF-067446846, 60 min, rep 4 -riboseq; Homo sapiens; OTHER GSM2476015: 1.5 ?M PF-067446846, 60 min, rep 4 -riboseq; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956371 Illumina HiSeq 2500 57588015 1357395400 SRR5227306 57588015 2936988765 SRP098789
SRP098789 SRX2536420 GSM2476014: vehicle, 60 min rep 3; Homo sapiens; OTHER GSM2476014: vehicle, 60 min rep 3; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956370 Illumina HiSeq 2000 48405034 1530784033 SRR5227305 48405034 2420251700 SRP098789
SRP098789 SRX2536419 GSM2476013: vehicle, 60 min rep 2; Homo sapiens; OTHER GSM2476013: vehicle, 60 min rep 2; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956369 Illumina HiSeq 2000 47139057 1489018603 SRR5227304 47139057 2356952850 SRP098789
SRP098789 SRX2536418 GSM2476012: vehicle, 60 min rep 1; Homo sapiens; OTHER GSM2476012: vehicle, 60 min rep 1; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956368 Illumina HiSeq 2000 50956178 1495757884 SRR5227303 50956178 2547808900 SRP098789
SRP098789 SRX2536417 GSM2476011: 0.3 ?M PF-067446846, 60 min, rep 3; Homo sapiens; OTHER GSM2476011: 0.3 ?M PF-067446846, 60 min, rep 3; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956367 Illumina HiSeq 2000 44258180 1404548468 SRR5227302 44258180 2212909000 SRP098789
SRP098789 SRX2536416 GSM2476010: 0.3 ?M PF-067446846, 60 min, rep 2; Homo sapiens; OTHER GSM2476010: 0.3 ?M PF-067446846, 60 min, rep 2; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956366 Illumina HiSeq 2000 49129512 1536091510 SRR5227301 49129512 2456475600 SRP098789
SRP098789 SRX2536415 GSM2476009: 0.3 ?M PF-067446846, 60 min, rep 1; Homo sapiens; OTHER GSM2476009: 0.3 ?M PF-067446846, 60 min, rep 1; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956365 Illumina HiSeq 2000 30043362 903983724 SRR5227300 30043362 1502168100 SRP098789
SRP098789 SRX2536414 GSM2476008: 1.5 ?M PF-067446846, 60 min, rep 3; Homo sapiens; OTHER GSM2476008: 1.5 ?M PF-067446846, 60 min, rep 3; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956364 Illumina HiSeq 2000 48766213 1530350854 SRR5227299 48766213 2438310650 SRP098789
SRP098789 SRX2536413 GSM2476007: 1.5 ?M PF-067446846, 60 min, rep 2; Homo sapiens; OTHER GSM2476007: 1.5 ?M PF-067446846, 60 min, rep 2; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956363 Illumina HiSeq 2000 49334392 1475414353 SRR5227298 49334392 2466719600 SRP098789
SRP098789 SRX2536412 GSM2476006: 1.5 ?M PF-067446846, 60 min, rep 1; Homo sapiens; OTHER GSM2476006: 1.5 ?M PF-067446846, 60 min, rep 1; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956362 Illumina HiSeq 2000 60381365 1801283052 SRR5227297 60381365 3019068250 SRP098789
SRP098789 SRX2536411 GSM2476005: vehicle, 10 min rep 3; Homo sapiens; OTHER GSM2476005: vehicle, 10 min rep 3; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956361 Illumina HiSeq 2000 52737784 1644829192 SRR5227296 52737784 2636889200 SRP098789
SRP098789 SRX2536410 GSM2476004: vehicle, 10 min rep 2; Homo sapiens; OTHER GSM2476004: vehicle, 10 min rep 2; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956360 Illumina HiSeq 2000 46137148 1455541408 SRR5227295 46137148 2306857400 SRP098789
SRP098789 SRX2536409 GSM2476003: vehicle, 10 min rep 1; Homo sapiens; OTHER GSM2476003: vehicle, 10 min rep 1; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956359 Illumina HiSeq 2000 76002122 1552821132 SRR5227294 76002122 3800106100 SRP098789
SRP098789 SRX2536408 GSM2476002: 0.3 ?M PF-067446846, 10 min, rep 3; Homo sapiens; OTHER GSM2476002: 0.3 ?M PF-067446846, 10 min, rep 3; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956358 Illumina HiSeq 2000 42709138 1338829352 SRR5227293 42709138 2135456900 SRP098789
SRP098789 SRX2536407 GSM2476001: 0.3 ?M PF-067446846, 10 min, rep 2; Homo sapiens; OTHER GSM2476001: 0.3 ?M PF-067446846, 10 min, rep 2; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956357 Illumina HiSeq 2000 60552437 1875910244 SRR5227292 60552437 3027621850 SRP098789
SRP098789 SRX2536406 GSM2476000: 0.3 ?M PF-067446846, 10 min, rep 1; Homo sapiens; OTHER GSM2476000: 0.3 ?M PF-067446846, 10 min, rep 1; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956356 Illumina HiSeq 2000 41143319 843881081 SRR5227291 41143319 2057165950 SRP098789
SRP098789 SRX2536405 GSM2475999: 1.5 ?M PF-067446846, 10 min, rep 3; Homo sapiens; OTHER GSM2475999: 1.5 ?M PF-067446846, 10 min, rep 3; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956355 Illumina HiSeq 2000 40462973 1287284933 SRR5227290 40462973 2023148650 SRP098789
SRP098789 SRX2536404 GSM2475998: 1.5 ?M PF-067446846, 10 min, rep 2; Homo sapiens; OTHER GSM2475998: 1.5 ?M PF-067446846, 10 min, rep 2; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956354 Illumina HiSeq 2000 41657461 1360366732 SRR5227289 41657461 2082873050 SRP098789
SRP098789 SRX2536403 GSM2475997: 1.5 ?M PF-067446846, 10 min, rep 1; Homo sapiens; OTHER GSM2475997: 1.5 ?M PF-067446846, 10 min, rep 1; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956353 Illumina HiSeq 2000 42082855 916745706 SRR5227288 42082855 2104142750 SRP098789
Convert GSE to SRP¶
$ pysradb srp-to-srx SRP098789
from pysradb.sraweb import SRAweb
client = SRAweb()
df = client.srp-to-srx("SRP098789")
df
Output:
study_accession experiment_accession experiment_title experiment_desc organism_taxid organism_name library_strategy library_source library_selection sample_accession sample_title instrument total_spots total_size run_accession run_total_spots run_total_bases study_accesssion
SRP098789 SRX2536428 GSM2476022: vehicle, 60 min, rep 5-mRNAseq; Homo sapiens; RNA-Seq GSM2476022: vehicle, 60 min, rep 5-mRNAseq; Homo sapiens; RNA-Seq 9606 Homo sapiens RNA-Seq TRANSCRIPTOMIC cDNA SRS1956378 Illumina HiSeq 2500 69422931 1545681856 SRR5227313 69422931 3540569481 SRP098789
SRP098789 SRX2536427 GSM2476021: PF-06446846, 60 min, rep 5 -mRNA-seq; Homo sapiens; OTHER GSM2476021: PF-06446846, 60 min, rep 5 -mRNA-seq; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956377 Illumina HiSeq 2500 58065134 1302369810 SRR5227312 58065134 2961321834 SRP098789
SRP098789 SRX2536426 GSM2476020: vehicle, 60 min, rep 4-mRNAseq; Homo sapiens; RNA-Seq GSM2476020: vehicle, 60 min, rep 4-mRNAseq; Homo sapiens; RNA-Seq 9606 Homo sapiens RNA-Seq TRANSCRIPTOMIC cDNA SRS1956376 Illumina HiSeq 2500 63720205 1416818619 SRR5227311 63720205 3249730455 SRP098789
SRP098789 SRX2536425 GSM2476019: PF-06446846, 60 min, rep 4 -mRNA-seq; Homo sapiens; OTHER GSM2476019: PF-06446846, 60 min, rep 4 -mRNA-seq; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956375 Illumina HiSeq 2500 66363585 1482728577 SRR5227310 66363585 3384542835 SRP098789
SRP098789 SRX2536424 GSM2476018: vehicle, 60 min, rep 5-Ribo-seq; Homo sapiens; RNA-Seq GSM2476018: vehicle, 60 min, rep 5-Ribo-seq; Homo sapiens; RNA-Seq 9606 Homo sapiens RNA-Seq TRANSCRIPTOMIC cDNA SRS1956374 Illumina HiSeq 2500 40062613 904488287 SRR5227309 40062613 2043193263 SRP098789
SRP098789 SRX2536423 GSM2476017: 1.5 ?M PF-067446846, 60 min, rep 5 -riboseq; Homo sapiens; OTHER GSM2476017: 1.5 ?M PF-067446846, 60 min, rep 5 -riboseq; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956373 Illumina HiSeq 2500 65591217 1499668100 SRR5227308 65591217 3345152067 SRP098789
SRP098789 SRX2536422 GSM2476016: Vehicle, 60 min, rep 4-ribo-seq; Homo sapiens; RNA-Seq GSM2476016: Vehicle, 60 min, rep 4-ribo-seq; Homo sapiens; RNA-Seq 9606 Homo sapiens RNA-Seq TRANSCRIPTOMIC cDNA SRS1956372 Illumina HiSeq 2500 66480991 1564636133 SRR5227307 66480991 3390530541 SRP098789
SRP098789 SRX2536421 GSM2476015: 1.5 ?M PF-067446846, 60 min, rep 4 -riboseq; Homo sapiens; OTHER GSM2476015: 1.5 ?M PF-067446846, 60 min, rep 4 -riboseq; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956371 Illumina HiSeq 2500 57588015 1357395400 SRR5227306 57588015 2936988765 SRP098789
SRP098789 SRX2536420 GSM2476014: vehicle, 60 min rep 3; Homo sapiens; OTHER GSM2476014: vehicle, 60 min rep 3; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956370 Illumina HiSeq 2000 48405034 1530784033 SRR5227305 48405034 2420251700 SRP098789
SRP098789 SRX2536419 GSM2476013: vehicle, 60 min rep 2; Homo sapiens; OTHER GSM2476013: vehicle, 60 min rep 2; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956369 Illumina HiSeq 2000 47139057 1489018603 SRR5227304 47139057 2356952850 SRP098789
SRP098789 SRX2536418 GSM2476012: vehicle, 60 min rep 1; Homo sapiens; OTHER GSM2476012: vehicle, 60 min rep 1; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956368 Illumina HiSeq 2000 50956178 1495757884 SRR5227303 50956178 2547808900 SRP098789
SRP098789 SRX2536417 GSM2476011: 0.3 ?M PF-067446846, 60 min, rep 3; Homo sapiens; OTHER GSM2476011: 0.3 ?M PF-067446846, 60 min, rep 3; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956367 Illumina HiSeq 2000 44258180 1404548468 SRR5227302 44258180 2212909000 SRP098789
SRP098789 SRX2536416 GSM2476010: 0.3 ?M PF-067446846, 60 min, rep 2; Homo sapiens; OTHER GSM2476010: 0.3 ?M PF-067446846, 60 min, rep 2; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956366 Illumina HiSeq 2000 49129512 1536091510 SRR5227301 49129512 2456475600 SRP098789
SRP098789 SRX2536415 GSM2476009: 0.3 ?M PF-067446846, 60 min, rep 1; Homo sapiens; OTHER GSM2476009: 0.3 ?M PF-067446846, 60 min, rep 1; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956365 Illumina HiSeq 2000 30043362 903983724 SRR5227300 30043362 1502168100 SRP098789
SRP098789 SRX2536414 GSM2476008: 1.5 ?M PF-067446846, 60 min, rep 3; Homo sapiens; OTHER GSM2476008: 1.5 ?M PF-067446846, 60 min, rep 3; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956364 Illumina HiSeq 2000 48766213 1530350854 SRR5227299 48766213 2438310650 SRP098789
SRP098789 SRX2536413 GSM2476007: 1.5 ?M PF-067446846, 60 min, rep 2; Homo sapiens; OTHER GSM2476007: 1.5 ?M PF-067446846, 60 min, rep 2; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956363 Illumina HiSeq 2000 49334392 1475414353 SRR5227298 49334392 2466719600 SRP098789
SRP098789 SRX2536412 GSM2476006: 1.5 ?M PF-067446846, 60 min, rep 1; Homo sapiens; OTHER GSM2476006: 1.5 ?M PF-067446846, 60 min, rep 1; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956362 Illumina HiSeq 2000 60381365 1801283052 SRR5227297 60381365 3019068250 SRP098789
SRP098789 SRX2536411 GSM2476005: vehicle, 10 min rep 3; Homo sapiens; OTHER GSM2476005: vehicle, 10 min rep 3; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956361 Illumina HiSeq 2000 52737784 1644829192 SRR5227296 52737784 2636889200 SRP098789
SRP098789 SRX2536410 GSM2476004: vehicle, 10 min rep 2; Homo sapiens; OTHER GSM2476004: vehicle, 10 min rep 2; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956360 Illumina HiSeq 2000 46137148 1455541408 SRR5227295 46137148 2306857400 SRP098789
SRP098789 SRX2536409 GSM2476003: vehicle, 10 min rep 1; Homo sapiens; OTHER GSM2476003: vehicle, 10 min rep 1; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956359 Illumina HiSeq 2000 76002122 1552821132 SRR5227294 76002122 3800106100 SRP098789
SRP098789 SRX2536408 GSM2476002: 0.3 ?M PF-067446846, 10 min, rep 3; Homo sapiens; OTHER GSM2476002: 0.3 ?M PF-067446846, 10 min, rep 3; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956358 Illumina HiSeq 2000 42709138 1338829352 SRR5227293 42709138 2135456900 SRP098789
SRP098789 SRX2536407 GSM2476001: 0.3 ?M PF-067446846, 10 min, rep 2; Homo sapiens; OTHER GSM2476001: 0.3 ?M PF-067446846, 10 min, rep 2; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956357 Illumina HiSeq 2000 60552437 1875910244 SRR5227292 60552437 3027621850 SRP098789
SRP098789 SRX2536406 GSM2476000: 0.3 ?M PF-067446846, 10 min, rep 1; Homo sapiens; OTHER GSM2476000: 0.3 ?M PF-067446846, 10 min, rep 1; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956356 Illumina HiSeq 2000 41143319 843881081 SRR5227291 41143319 2057165950 SRP098789
SRP098789 SRX2536405 GSM2475999: 1.5 ?M PF-067446846, 10 min, rep 3; Homo sapiens; OTHER GSM2475999: 1.5 ?M PF-067446846, 10 min, rep 3; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956355 Illumina HiSeq 2000 40462973 1287284933 SRR5227290 40462973 2023148650 SRP098789
SRP098789 SRX2536404 GSM2475998: 1.5 ?M PF-067446846, 10 min, rep 2; Homo sapiens; OTHER GSM2475998: 1.5 ?M PF-067446846, 10 min, rep 2; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956354 Illumina HiSeq 2000 41657461 1360366732 SRR5227289 41657461 2082873050 SRP098789
SRP098789 SRX2536403 GSM2475997: 1.5 ?M PF-067446846, 10 min, rep 1; Homo sapiens; OTHER GSM2475997: 1.5 ?M PF-067446846, 10 min, rep 1; Homo sapiens; OTHER 9606 Homo sapiens OTHER TRANSCRIPTOMIC other SRS1956353 Illumina HiSeq 2000 42082855 916745706 SRR5227288 42082855 2104142750 SRP098789
Downloading sequencing data¶
pysradb can alse be used to download either .fastq or .sra
filesboth from ENA and SRA.
Downloading via accession number¶
$ pysradb download SRP098789
from pysradb.sraweb import SRAweb
client = SRAweb()
client.download("SRP098789")
It is also possible to pipe the dataframe from [metadata]{.title-ref} or [search]{.title-ref} to download, after filtering the dataframe entries:
$ pysradb metadata SRP276671 --detailed | pysradb download
from pysradb.sraweb import SRAweb
client = SRAweb()
df = client.sra_metadata('SRP016501', detailed=True)
client.download(df=df)
Ultrafast fastq downloads¶
With
aspera-client
installed, pysradb canan perform ultra fast downloads:
To download all original fastqs with [aspera-client]{.title-ref} installed utilizing 8 threads:
$ pysradb download -t 8 --use_ascp -p SRP002605
from pysradb.sraweb import SRAweb
client = SRAweb()
client.download("SRP098789", use_ascp=True, threads=8)
Refer to the notebook for (shallow) time benchmarks.
Search¶
Retrieving metadata by accession number¶
$ pysradb metadata SRP276671
from pysradb.sraweb import SRAweb
client = SRAweb()
df = client.sra_metadata('SRP016501')
df
Output:
study_accession experiment_accession experiment_title experiment_desc organism_taxid organism_name library_strategy library_source library_selection sample_accession sample_title instrument total_spots total_size run_accession run_total_spots run_total_bases
SRP276671 SRX8978626 hCov-19/Canada/ON/VIDO01/2020 (EPI ISL 413015) hCov-19/Canada/ON/VIDO01/2020 (EPI ISL 413015) 2697049 Severe acute respiratory syndrome coronavirus 2 WGS GENOMIC RT-PCR SRS7233795 MinION 96202 79690689 SRR12486810 96202 86575096
SRP276671 SRX8909137 hCoV-19/Canada/ON-VIDO-01/2020 (EPI_ISL_425177) hCoV-19/Canada/ON-VIDO-01/2020 (EPI_ISL_425177) 2697049 Severe acute respiratory syndrome coronavirus 2 WGS GENOMIC RT-PCR SRS7166526 Illumina MiSeq 866225 173474986 SRR12412952 866225 338457239
::: note ::: title Note :::
[pysradb]{.title-ref}, when used in python, returns the retrieved metadata as a [pandas]{.title-ref} DataFrame, with all regular select/query operations available through [pandas]{.title-ref}. :::
For more detailed metadata (including download URLs), we can include the [detailed]{.title-ref} flag:
$ pysradb metadata SRP276671 --detailed
from pysradb.sraweb import SRAweb
client = SRAweb()
df = client.sra_metadata('SRP016501', detailed=True)
df
Output:
study_accession experiment_accession experiment_title experiment_desc organism_taxid organism_name library_strategy library_source library_selection sample_accession sample_title instrument total_spots total_size run_accession run_total_spots run_total_bases run_alias sra_url_alt sra_url experiment_alias isolate collected_by collection_date geo_loc_name host host_disease isolation_source lat_lon host_sex host_subject_id passage_history BioSampleModel sra_url_alt1 sra_url_alt2 sra_url_alt3 ena_fastq_http ena_fastq_ftp
SRP276671 SRX8978626 hCov-19/Canada/ON/VIDO01/2020 (EPI ISL 413015) hCov-19/Canada/ON/VIDO01/2020 (EPI ISL 413015) 2697049 Severe acute respiratory syndrome coronavirus 2 WGS GENOMIC RT-PCR SRS7233795 N/A MinION 96202 79690689 SRR12486810 96202 86575096 VIDO-01.tar.gz https://sra-download.ncbi.nlm.nih.gov/traces/sra1/SRZ/012486/SRR12486810/VIDO-01.tar.gz https://sra-download.ncbi.nlm.nih.gov/traces/sra78/SRR/012194/SRR12486810 N/A ON-VIDO-01-P3 Public Health Ontario 2020-01-23 Canada: Ontario Homo sapiens COVID-19 missing missing male VIDO-01 Vero E6 P3 Pathogen.cl N/A N/A N/A http://ftp.sra.ebi.ac.uk/vol1/fastq/SRR124/010/SRR12486810/SRR12486810_1.fastq.gz era-fasp@fasp.sra.ebi.ac.uk:vol1/fastq/SRR124/010/SRR12486810/SRR12486810_1.fastq.gz
SRP276671 SRX8909137 hCoV-19/Canada/ON-VIDO-01/2020 (EPI_ISL_425177) hCoV-19/Canada/ON-VIDO-01/2020 (EPI_ISL_425177) 2697049 Severe acute respiratory syndrome coronavirus 2 WGS GENOMIC RT-PCR SRS7166526 N/A Illumina MiSeq 866225 173474986 SRR12412952 866225 338457239 SP-2_R1.fastq.gz N/A https://sra-download.ncbi.nlm.nih.gov/traces/sra48/SRR/012122/SRR12412952 N/A ON-VIDO-01-P2 Public Health Ontario 2020-01-23 Canada: Ontario Homo sapiens COVID-19 missing missing male VIDO-01 Vero E6 P2 Pathogen.cl gs://sra-pub-src-10/SRR12412952/SP-2_R2.fastq.1 s3://sra-pub-src-10/SRR12412952/SP-2_R2.fastq.1 https://sra-pub-sars-cov2.s3.amazonaws.com/sra-src/SRR12412952/SP-2_R2.fastq.1 http://ftp.sra.ebi.ac.uk/vol1/fastq/SRR124/052/SRR12412952/SRR12412952.fastq.gz era-fasp@fasp.sra.ebi.ac.uk:vol1/fastq/SRR124/052/SRR12412952/SRR12412952.fastq.gz
Searching SRA/ENA databases and retrieving metadata¶
Let’s take for example we are interested in coronavirus sequences published on Short Reads Archive (SRA) in the first week of August 2020.
$ pysradb search -q coronavirus --publication-date 01-08-2020:07-08-2020 --max 10000
from pysradb.search import SraSearch
instance = SraSearch(return_max=10000, query="coronavirus", publication_date="01-08-2020:07-08-2020")
instance.search()
instance.get_df()
Output (showing only the first 10 entries): :
study_accession experiment_accession experiment_title sample_taxon_id sample_scientific_name experiment_library_strategy experiment_library_source experiment_library_selection sample_accession sample_alias experiment_instrument_model pool_member_spots run_1_size run_1_accession run_1_total_spots run_1_total_bases pmid
SRP270658 SRX8679965 GSM4658808: SARS-CoV-2-infected 24h 3; Chlorocebus sabaeus; Severe acute respiratory syndrome coronavirus 2; RNA-Seq 60711 Chlorocebus sabaeus RNA-Seq TRANSCRIPTOMIC cDNA SRS6959042 GSM4658808 NextSeq 500 104223040 9743267247 SRR12164500 104223040 31475358080 11295714
SRP270658 SRX8679964 GSM4658807: SARS-CoV-2-infected 24h 2; Chlorocebus sabaeus; Severe acute respiratory syndrome coronavirus 2; RNA-Seq 60711 Chlorocebus sabaeus RNA-Seq TRANSCRIPTOMIC cDNA SRS6959041 GSM4658807 NextSeq 500 92813819 8703506222 SRR12164499 92813819 28029773338 11295713
SRP253798 SRX8677889 Severe acute respiratory syndrome coronavirus 2 2697049 Severe acute respiratory syndrome coronavirus 2 AMPLICON VIRAL RNA PCR SRS6956975 hCoV-19/Australia/VIC1898/2020 NextSeq 500 456828 51422072 SRR12162149 456828 130280958 11292876
SRP253798 SRX8677888 Severe acute respiratory syndrome coronavirus 2 2697049 Severe acute respiratory syndrome coronavirus 2 AMPLICON VIRAL RNA PCR SRS6956974 hCoV-19/Australia/VIC1886/2020 NextSeq 500 268832 29923966 SRR12162150 268832 75885223 11292875
SRP253798 SRX8677887 Severe acute respiratory syndrome coronavirus 2 2697049 Severe acute respiratory syndrome coronavirus 2 AMPLICON VIRAL RNA PCR SRS6956973 hCoV-19/Australia/VIC1890/2020 NextSeq 500 483526 54629557 SRR12162151 483526 139019404 11292874
SRP253798 SRX8677886 Severe acute respiratory syndrome coronavirus 2 2697049 Severe acute respiratory syndrome coronavirus 2 AMPLICON VIRAL RNA PCR SRS6956971 hCoV-19/Australia/VIC1888/2020 NextSeq 500 473895 53675126 SRR12162152 473895 136058655 11292873
SRP253798 SRX8677885 Severe acute respiratory syndrome coronavirus 2 2697049 Severe acute respiratory syndrome coronavirus 2 AMPLICON VIRAL RNA PCR SRS6956972 hCoV-19/Australia/VIC1891/2020 NextSeq 500 482373 53331905 SRR12162153 482373 135769259 11292872
SRP253798 SRX8677884 Severe acute respiratory syndrome coronavirus 2 2697049 Severe acute respiratory syndrome coronavirus 2 AMPLICON VIRAL RNA PCR SRS6956970 hCoV-19/Australia/VIC1816/2020 NextSeq 550 357052 41111134 SRR12162154 357052 103693201 11292871
SRP253798 SRX8677883 Severe acute respiratory syndrome coronavirus 2 2697049 Severe acute respiratory syndrome coronavirus 2 AMPLICON VIRAL RNA PCR SRS6956969 hCoV-19/Australia/VIC1815/2020 NextSeq 550 307106 35306959 SRR12162155 307106 89866234 11292870
SRP253798 SRX8677882 Severe acute respiratory syndrome coronavirus 2 2697049 Severe acute respiratory syndrome coronavirus 2 AMPLICON VIRAL RNA PCR SRS6956968 hCoV-19/Australia/VIC1814/2020 NextSeq 550 353704 40652239 SRR12162156 353704 103366580 11292869
To query European Nucleotide Archive (ENA) instead:
$ pysradb search --db ena -q coronavirus --publication-date 01-08-2020:07-08-2020 --max 10000
from pysradb.search import EnaSearch
instance = EnaSearch(return_max=10000, query="coronavirus", publication_date="01-08-2020:07-08-2020")
instance.search()
instance.get_df()
Output (showing only the first 10 entries): :
study_accession experiment_accession experiment_title description tax_id scientific_name library_strategylibrary_source library_selection sample_accession sample_title instrument_model run_accession read_count base_count
PRJEB12126 ERX1264364 Illumina HiSeq 2000 sequencing; Analysis of coronavirus and infected host-cell gene expression through RNA sequencing and ribosome profiling Illumina HiSeq 2000 sequencing; Analysis of coronavirus and infected host-cell gene expression through RNA sequencing and ribosome profiling 10090 Mus musculus OTHER TRANSCRIPTOMIC other SAMEA3708907 Sample 1 Illumina HiSeq 2000 ERR1190989 38883498 1161289538
PRJEB12126 ERX1264365 Illumina HiSeq 2000 sequencing; Analysis of coronavirus and infected host-cell gene expression through RNA sequencing and ribosome profiling Illumina HiSeq 2000 sequencing; Analysis of coronavirus and infected host-cell gene expression through RNA sequencing and ribosome profiling 10090 Mus musculus OTHER TRANSCRIPTOMIC other SAMEA3708908 Sample 10 Illumina HiSeq 2000 ERR1190990 55544297 1779600908
PRJEB12126 ERX1264366 Illumina HiSeq 2000 sequencing; Analysis of coronavirus and infected host-cell gene expression through RNA sequencing and ribosome profiling Illumina HiSeq 2000 sequencing; Analysis of coronavirus and infected host-cell gene expression through RNA sequencing and ribosome profiling 10090 Mus musculus OTHER TRANSCRIPTOMIC other SAMEA3708909 Sample 11 Illumina HiSeq 2000 ERR1190991 54474851 1713994365
PRJEB12126 ERX1264367 Illumina HiSeq 2000 sequencing; Analysis of coronavirus and infected host-cell gene expression through RNA sequencing and ribosome profiling Illumina HiSeq 2000 sequencing; Analysis of coronavirus and infected host-cell gene expression through RNA sequencing and ribosome profiling 10090 Mus musculus OTHER TRANSCRIPTOMIC other SAMEA3708910 Sample 12 Illumina HiSeq 2000 ERR1190992 78497711 2489092061
PRJEB12126 ERX1264368 Illumina HiSeq 2000 sequencing; Analysis of coronavirus and infected host-cell gene expression through RNA sequencing and ribosome profiling Illumina HiSeq 2000 sequencing; Analysis of coronavirus and infected host-cell gene expression through RNA sequencing and ribosome profiling 10090 Mus musculus RNA-Seq TRANSCRIPTOMIC RANDOM SAMEA3708911 Sample 13 Illumina HiSeq 2000 ERR1190993 84955423 2627276298
PRJEB12126 ERX1264369 Illumina HiSeq 2000 sequencing; Analysis of coronavirus and infected host-cell gene expression through RNA sequencing and ribosome profiling Illumina HiSeq 2000 sequencing; Analysis of coronavirus and infected host-cell gene expression through RNA sequencing and ribosome profiling 10090 Mus musculus RNA-Seq TRANSCRIPTOMIC RANDOM SAMEA3708912 Sample 14 Illumina HiSeq 2000 ERR1190994 75097651 2293097872
PRJEB12126 ERX1264370 Illumina HiSeq 2000 sequencing; Analysis of coronavirus and infected host-cell gene expression through RNA sequencing and ribosome profiling Illumina HiSeq 2000 sequencing; Analysis of coronavirus and infected host-cell gene expression through RNA sequencing and ribosome profiling 10090 Mus musculus RNA-Seq TRANSCRIPTOMIC RANDOM SAMEA3708913 Sample 15 Illumina HiSeq 2000 ERR1190995 67177553 2060926619
PRJEB12126 ERX1264371 Illumina HiSeq 2000 sequencing; Analysis of coronavirus and infected host-cell gene expression through RNA sequencing and ribosome profiling Illumina HiSeq 2000 sequencing; Analysis of coronavirus and infected host-cell gene expression through RNA sequencing and ribosome profiling 10090 Mus musculus RNA-Seq TRANSCRIPTOMIC RANDOM SAMEA3708914 Sample 16 Illumina HiSeq 2000 ERR1190996 62940694 2061757111
PRJEB12126 ERX1264372 Illumina HiSeq 2000 sequencing; Analysis of coronavirus and infected host-cell gene expression through RNA sequencing and ribosome profiling Illumina HiSeq 2000 sequencing; Analysis of coronavirus and infected host-cell gene expression through RNA sequencing and ribosome profiling 10090 Mus musculus RNA-Seq TRANSCRIPTOMIC RANDOM SAMEA3708915 Sample 17 Illumina HiSeq 2000 ERR1190997 80591061 2475034240
PRJEB12126 ERX1264373 Illumina HiSeq 2000 sequencing; Analysis of coronavirus and infected host-cell gene expression through RNA sequencing and ribosome profiling Illumina HiSeq 2000 sequencing; Analysis of coronavirus and infected host-cell gene expression through RNA sequencing and ribosome profiling 10090 Mus musculus RNA-Seq TRANSCRIPTOMIC RANDOM SAMEA3708916 Sample 18 Illumina HiSeq 2000 ERR1190998 68575621 2149386138
If the number of returned entries is large, it might be troublesome to filter through the metadata to find any information of interest. As a starting point, we can use the search feature to generate summary statistics and graphs for the search result:
Statistics¶
$ pysradb search --db ena --organism "Severe acute respiratory syndrome coronavirus 2" --max 10000 -s
from pysradb.search import EnaSearch
instance = EnaSearch(return_max=10000, organism="Severe acute respiratory syndrome coronavirus 2")
instance.search()
instance.show_result_statistics()
Output:
Statistics for the search query:
=================================
Number of unique studies: 7
Number of unique experiments: 10000
Number of unique runs: 10000
Number of unique samples: 9797
Mean base count of samples: 238380171.626
Median base count of samples: 164470138.000
Sample base count standard deviation: 261654776.053
Date range:
2020-04: 1299
2020-05: 2518
2020-06: 6181
2020-07: 2
Organisms:
Severe acute respiratory syndrome coronavirus 2: 10000
Platform:
ILLUMINA: 5175
OXFORD_NANOPORE: 4825
Library strategy:
AMPLICON: 9789
RNA-Seq: 1
Targeted-Capture: 202
WGS: 8
Library source:
GENOMIC: 8
METATRANSCRIPTOMIC: 1
TRANSCRIPTOMIC: 1
VIRAL RNA: 9990
Library selection:
PCR: 9789
RANDOM: 9
other: 202
Library layout:
PAIRED: 5059
SINGLE: 4941
Plotting¶
$ pysradb search --db ena -q e --max 500000 -g
from pysradb.search import EnaSearch
instance = EnaSearch(return_max=500000, "e")
instance.search()
instance.visualise_results()
Output: Graphs generated will automatically be saved under [./search_plots/]{.title-ref}. Optionally, graphs can be shown in python by including the argument [show=True]{.title-ref} Here are some of the available graphs that will be generated:
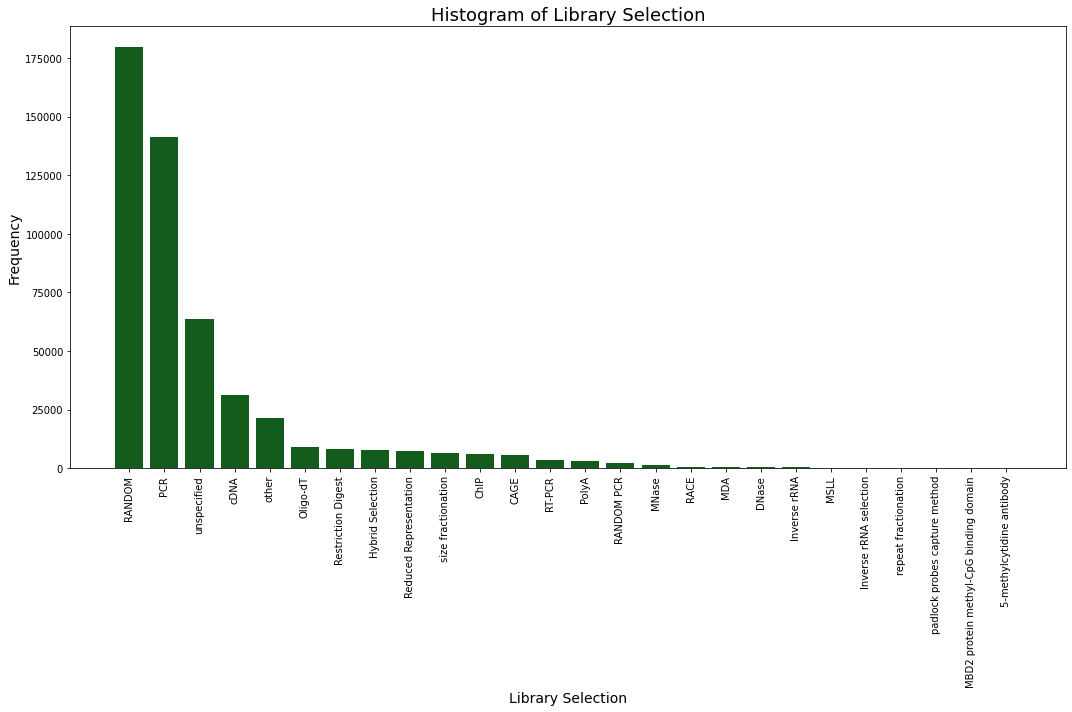 {width=”600px”}
{width=”600px”}
 {width=”600px”}
{width=”600px”}
 {width=”600px”}
{width=”600px”}
List of possible [pysradb]{.title-ref} operations¶
$ pysradb
usage: pysradb [-h] [--version] [--citation]
{metadb,metadata,download,search,gse-to-gsm,gse-to-srp,gsm-to-gse,gsm-to-srp,gsm-to-srr,gsm-to-srs,gsm-to-srx,srp-to-gse,srp-to-srr,srp-to-srs,srp-to-srx,srr-to-gsm,srr-to-srp,srr-to-srs,srr-to-srx,srs-to-gsm,srs-to-srx,srx-to-srp,srx-to-srr,srx-to-srs}
...
pysradb: Query NGS metadata and data from NCBI Sequence Read Archive.
Citation: 10.12688/f1000research.18676.1
optional arguments:
-h, --help show this help message and exit
--version show program's version number and exit
--citation how to cite
subcommands:
{metadb,metadata,download,search,gse-to-gsm,gse-to-srp,gsm-to-gse,gsm-to-srp,gsm-to-srr,gsm-to-srs,gsm-to-srx,srp-to-gse,srp-to-srr,srp-to-srs,srp-to-srx,srr-to-gsm,srr-to-srp,srr-to-srs,srr-to-srx,srs-to-gsm,srs-to-srx,srx-to-srp,srx-to-srr,srx-to-srs}
metadata Fetch metadata for SRA project (SRPnnnn)
download Download SRA project (SRPnnnn)
search Search SRA/ENA for matching text
gse-to-gsm Get GSM for a GSE
gse-to-srp Get SRP for a GSE
gsm-to-gse Get GSE for a GSM
gsm-to-srp Get SRP for a GSM
gsm-to-srr Get SRR for a GSM
gsm-to-srs Get SRS for a GSM
gsm-to-srx Get SRX for a GSM
srp-to-gse Get GSE for a SRP
srp-to-srr Get SRR for a SRP
srp-to-srs Get SRS for a SRP
srp-to-srx Get SRX for a SRP
srr-to-gsm Get GSM for a SRR
srr-to-srp Get SRP for a SRR
srr-to-srs Get SRS for a SRR
srr-to-srx Get SRX for a SRR
srs-to-gsm Get GSM for a SRS
srs-to-srx Get SRX for a SRS
srx-to-srp Get SRP for a SRX
srx-to-srr Get SRR for a SRX
srx-to-srs Get SRS for a SRX